Genomic data in the All of Us Research Program
- PMID: 38374255
- PMCID: PMC10937371
- DOI: 10.1038/s41586-023-06957-x
Genomic data in the All of Us Research Program
Abstract
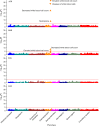
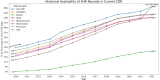
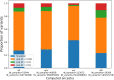
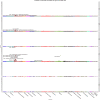
Comprehensively mapping the genetic basis of human disease across diverse individuals is a long-standing goal for the field of human genetics1-4. The All of Us Research Program is a longitudinal cohort study aiming to enrol a diverse group of at least one million individuals across the USA to accelerate biomedical research and improve human health5,6. Here we describe the programme's genomics data release of 245,388 clinical-grade genome sequences. This resource is unique in its diversity as 77% of participants are from communities that are historically under-represented in biomedical research and 46% are individuals from under-represented racial and ethnic minorities. All of Us identified more than 1 billion genetic variants, including more than 275 million previously unreported genetic variants, more than 3.9 million of which had coding consequences. Leveraging linkage between genomic data and the longitudinal electronic health record, we evaluated 3,724 genetic variants associated with 117 diseases and found high replication rates across both participants of European ancestry and participants of African ancestry. Summary-level data are publicly available, and individual-level data can be accessed by researchers through the All of Us Researcher Workbench using a unique data passport model with a median time from initial researcher registration to data access of 29 hours. We anticipate that this diverse dataset will advance the promise of genomic medicine for all.
© 2024. The Author(s).
Conflict of interest statement
D.M.M., G.A.M., E.V., K.W., J.H., H.D., C.L.K., M.M., S.D., Z.K., E. Boerwinkle and R.A.G. declare that Baylor Genetics is a Baylor College of Medicine affiliate that derives revenue from genetic testing. Eric Venner is affiliated with Codified Genomics, a provider of genetic interpretation. E.E.E. is a scientific advisory board member of Variant Bio, Inc. A.G.B. is a scientific advisory board member of TenSixteen Bio. The remaining authors declare no competing interests.
Figures






References
MeSH terms
Grants and funding
- OT2 OD038122/OD/NIH HHS/United States
- OT2 OD026556/OD/NIH HHS/United States
- U2C OD023196/OD/NIH HHS/United States
- OT2 OD026551/OD/NIH HHS/United States
- OT2 OD026552/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- OT2 OD025337/OD/NIH HHS/United States
- OT2 OD026550/OD/NIH HHS/United States
- OT2 OD026553/OD/NIH HHS/United States
- OT2 OD023205/OD/NIH HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- OT2 OD026554/OD/NIH HHS/United States
- U24 OD023163/OD/NIH HHS/United States
- OT2 OD023206/OD/NIH HHS/United States
- OT2 OD002748/OD/NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- OT2 OD026548/OD/NIH HHS/United States
- OT2 OD035404/OD/NIH HHS/United States
- OT2 OD025315/OD/NIH HHS/United States
- U24 OD023121/OD/NIH HHS/United States
- P50 HD103538/HD/NICHD NIH HHS/United States
- OT2 OD025277/OD/NIH HHS/United States
- RM1 HG012334/HG/NHGRI NIH HHS/United States
- DP2 OD002750/OD/NIH HHS/United States
- OT2 OD002750/OD/NIH HHS/United States
- OT2 OD026555/OD/NIH HHS/United States
- OT2 OD026557/OD/NIH HHS/United States
- OT2 OD002751/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

