A terminal metabolite of niacin promotes vascular inflammation and contributes to cardiovascular disease risk
- PMID: 38374343
- PMCID: PMC11841810
- DOI: 10.1038/s41591-023-02793-8
A terminal metabolite of niacin promotes vascular inflammation and contributes to cardiovascular disease risk
Erratum in
-
Publisher Correction: A terminal metabolite of niacin promotes vascular inflammation and contributes to cardiovascular disease risk.Nat Med. 2024 Jun;30(6):1791. doi: 10.1038/s41591-024-02899-7. Nat Med. 2024. PMID: 38448791 No abstract available.
Abstract
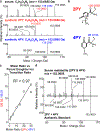
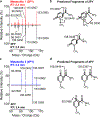
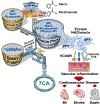
Despite intensive preventive cardiovascular disease (CVD) efforts, substantial residual CVD risk remains even for individuals receiving all guideline-recommended interventions. Niacin is an essential micronutrient fortified in food staples, but its role in CVD is not well understood. In this study, untargeted metabolomics analysis of fasting plasma from stable cardiac patients in a prospective discovery cohort (n = 1,162 total, n = 422 females) suggested that niacin metabolism was associated with incident major adverse cardiovascular events (MACE). Serum levels of the terminal metabolites of excess niacin, N1-methyl-2-pyridone-5-carboxamide (2PY) and N1-methyl-4-pyridone-3-carboxamide (4PY), were associated with increased 3-year MACE risk in two validation cohorts (US n = 2,331 total, n = 774 females; European n = 832 total, n = 249 females) (adjusted hazard ratio (HR) (95% confidence interval) for 2PY: 1.64 (1.10-2.42) and 2.02 (1.29-3.18), respectively; for 4PY: 1.89 (1.26-2.84) and 1.99 (1.26-3.14), respectively). Phenome-wide association analysis of the genetic variant rs10496731, which was significantly associated with both 2PY and 4PY levels, revealed an association of this variant with levels of soluble vascular adhesion molecule 1 (sVCAM-1). Further meta-analysis confirmed association of rs10496731 with sVCAM-1 (n = 106,000 total, n = 53,075 females, P = 3.6 × 10-18). Moreover, sVCAM-1 levels were significantly correlated with both 2PY and 4PY in a validation cohort (n = 974 total, n = 333 females) (2PY: rho = 0.13, P = 7.7 × 10-5; 4PY: rho = 0.18, P = 1.1 × 10-8). Lastly, treatment with physiological levels of 4PY, but not its structural isomer 2PY, induced expression of VCAM-1 and leukocyte adherence to vascular endothelium in mice. Collectively, these results indicate that the terminal breakdown products of excess niacin, 2PY and 4PY, are both associated with residual CVD risk. They also suggest an inflammation-dependent mechanism underlying the clinical association between 4PY and MACE.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Declaration of Interests
Drs Hazen and Wang report being named as co-inventors on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. Drs Hazen and Wang also report having received royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Lab, a fully owned subsidiary of Quest Diagnostics, and Procter & Gamble. Dr. Hazen is a paid consultant for Zehna Therapeutics and Proctor & Gamble, and has received research funds from Zehna Therapeutics, Proctor & Gamble, Pfizer, and Roche Diagnostics. Dr. Tang is a consultant for Sequana Medical A.V., Cardiol Therapeutics, Genomics plc, Zehna Therapeutics, and Renovacor, and has received honoraria from Springer Nature for authorship/editorship and American Board of Internal Medicine for exam writing committee. All other authors declare no competing interests.
Figures










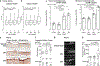
Comment in
-
Metabolic product of excess niacin is linked to increased risk of cardiovascular events.Nat Rev Cardiol. 2024 May;21(5):280. doi: 10.1038/s41569-024-01005-1. Nat Rev Cardiol. 2024. PMID: 38438588 No abstract available.
References
-
- D’Agostino RB et al. General cardiovascular risk profile for use in primary care: The Framingham heart study. Circulation 117, 743–753 (2008). - PubMed
-
- Ridker PM et al. Cardiovascular efficacy and safety of bococizumab in high-risk patients. N. Engl. J. Med. 376, 1527–1539 (2017). - PubMed
-
- Sabatine MS et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017). - PubMed
Methods-only References
-
- STROBE statement - Checklist of items that should be included in reports of observational studies (© STROBE Initiative). Int. J. Public Health 53, 3–4 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

