Nigral transcriptomic profiles in Engrailed-1 hemizygous mouse models of Parkinson's disease reveal upregulation of oxidative phosphorylation-related genes associated with delayed dopaminergic neurodegeneration
- PMID: 38374883
- PMCID: PMC10875038
- DOI: 10.3389/fnagi.2024.1337365
Nigral transcriptomic profiles in Engrailed-1 hemizygous mouse models of Parkinson's disease reveal upregulation of oxidative phosphorylation-related genes associated with delayed dopaminergic neurodegeneration
Abstract
Introduction: Parkinson's disease (PD) is the second most common neurodegenerative disorder, increasing both in terms of prevalence and incidence. To date, only symptomatic treatment is available, highlighting the need to increase knowledge on disease etiology in order to develop new therapeutic strategies. Hemizygosity for the gene Engrailed-1 (En1), encoding a conserved transcription factor essential for the programming, survival, and maintenance of midbrain dopaminergic neurons, leads to progressive nigrostriatal degeneration, motor impairment and depressive-like behavior in SwissOF1 (OF1-En1+/-). The neurodegenerative phenotype is, however, absent in C57Bl/6j (C57-En1+/-) mice. En1+/- mice are thus highly relevant tools to identify genetic factors underlying PD susceptibility.
Methods: Transcriptome profiles were defined by RNAseq in microdissected substantia nigra from 1-week old OF1, OF1- En1+/-, C57 and C57- En1+/- male mice. Differentially expressed genes (DEGs) were analyzed for functional enrichment. Neurodegeneration was assessed in 4- and 16-week old mice by histology.
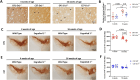
Results: Nigrostriatal neurodegeneration was manifested in OF1- En1+/- mice by increased dopaminergic striatal axonal swellings from 4 to 16 weeks and decreased number of dopaminergic neurons in the SNpc at 16 weeks compared to OF1. In contrast, C57- En1+/- mice had no significant increase in axonal swellings or cell loss in SNpc at 16 weeks. Transcriptomic analyses identified 198 DEGs between OF1- En1+/- and OF1 mice but only 52 DEGs between C57- En1+/- and C57 mice. Enrichment analysis of DEGs revealed that the neuroprotective phenotype of C57- En1+/- mice was associated with a higher expression of oxidative phosphorylation-related genes compared to both C57 and OF1- En1+/- mice.
Discussion: Our results suggest that increased expression of genes encoding mitochondrial proteins before the onset of neurodegeneration is associated with increased resistance to PD-like nigrostriatal neurodegeneration. This highlights the importance of genetic background in PD models, how different strains can be used to model clinical and sub-clinical pathologies and provides insights to gene expression mechanisms associated with PD susceptibility and progression.
Keywords: Parkinson’s disease; genetic susceptibility; mitochondria; neurodegeneration; substantia nigra; transcriptomics.
Copyright © 2024 Belfiori, Dueñas Rey, Ralbovszki, Jimenez-Ferrer, Fredlund, Balikai, Ahrén, Brolin and Swanberg.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Alvarez-Fischer D., Fuchs J., Castagner F. F., Stettler O., Massiani-Beaudoin O., Moya K. L., et al. . (2011). Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat. Neurosci. 14, 1260–1266. doi: 10.1038/nn.2916, PMID: Retraction in: Nat Neurosci. 2011 Oct;14(10):1221-2 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

