Genome-wide identification and characterization of TCP gene family in Dendrobium nobile and their role in perianth development
- PMID: 38375086
- PMCID: PMC10875090
- DOI: 10.3389/fpls.2024.1352119
Genome-wide identification and characterization of TCP gene family in Dendrobium nobile and their role in perianth development
Abstract
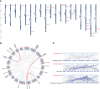
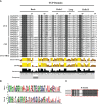
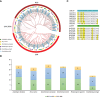
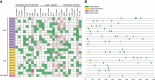
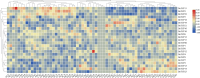
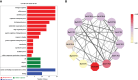
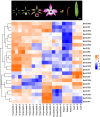
TCP is a widely distributed, essential plant transcription factor that regulates plant growth and development. An in-depth study of TCP genes in Dendrobium nobile, a crucial parent in genetic breeding and an excellent model material to explore perianth development in Dendrobium, has not been conducted. We identified 23 DnTCP genes unevenly distributed across 19 chromosomes and classified them as Class I PCF (12 members), Class II: CIN (10 members), and CYC/TB1 (1 member) based on the conserved domain and phylogenetic analysis. Most DnTCPs in the same subclade had similar gene and motif structures. Segmental duplication was the predominant duplication event for TCP genes, and no tandem duplication was observed. Seven genes in the CIN subclade had potential miR319 and -159 target sites. Cis-acting element analysis showed that most DnTCP genes contained many developmental stress-, light-, and phytohormone-responsive elements in their promoter regions. Distinct expression patterns were observed among the 23 DnTCP genes, suggesting that these genes have diverse regulatory roles at different stages of perianth development or in different organs. For instance, DnTCP4 and DnTCP18 play a role in early perianth development, and DnTCP5 and DnTCP10 are significantly expressed during late perianth development. DnTCP17, 20, 21, and 22 are the most likely to be involved in perianth and leaf development. DnTCP11 was significantly expressed in the gynandrium. Specially, MADS-specific binding sites were present in most DnTCP genes putative promoters, and two Class I DnTCPs were in the nucleus and interacted with each other or with the MADS-box. The interactions between TCP and the MADS-box have been described for the first time in orchids, which broadens our understanding of the regulatory network of TCP involved in perianth development in orchids.
Keywords: Dendrobium nobile; TCP gene family; expression pattern; perianth development; protein interaction.
Copyright © 2024 Wei, Yuan, Zheng, Zhou and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
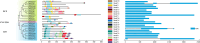


References
LinkOut - more resources
Full Text Sources

