Antiviral responses versus virus-induced cellular shutoff: a game of thrones between influenza A virus NS1 and SARS-CoV-2 Nsp1
- PMID: 38375361
- PMCID: PMC10875036
- DOI: 10.3389/fcimb.2024.1357866
Antiviral responses versus virus-induced cellular shutoff: a game of thrones between influenza A virus NS1 and SARS-CoV-2 Nsp1
Abstract
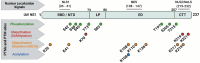
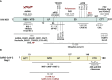
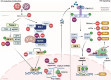
Following virus recognition of host cell receptors and viral particle/genome internalization, viruses replicate in the host via hijacking essential host cell machinery components to evade the provoked antiviral innate immunity against the invading pathogen. Respiratory viral infections are usually acute with the ability to activate pattern recognition receptors (PRRs) in/on host cells, resulting in the production and release of interferons (IFNs), proinflammatory cytokines, chemokines, and IFN-stimulated genes (ISGs) to reduce virus fitness and mitigate infection. Nevertheless, the game between viruses and the host is a complicated and dynamic process, in which they restrict each other via specific factors to maintain their own advantages and win this game. The primary role of the non-structural protein 1 (NS1 and Nsp1) of influenza A viruses (IAV) and the pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), respectively, is to control antiviral host-induced innate immune responses. This review provides a comprehensive overview of the genesis, spatial structure, viral and cellular interactors, and the mechanisms underlying the unique biological functions of IAV NS1 and SARS-CoV-2 Nsp1 in infected host cells. We also highlight the role of both non-structural proteins in modulating viral replication and pathogenicity. Eventually, and because of their important role during viral infection, we also describe their promising potential as targets for antiviral therapy and the development of live attenuated vaccines (LAV). Conclusively, both IAV NS1 and SARS-CoV-2 Nsp1 play an important role in virus-host interactions, viral replication, and pathogenesis, and pave the way to develop novel prophylactic and/or therapeutic interventions for the treatment of these important human respiratory viral pathogens.
Keywords: NS1; Nsp1; SARS-CoV-2; antivirals; influenza A virus; innate immunity; live attenuated vaccines; non-structural protein 1.
Copyright © 2024 Khalil, Nogales, Martínez-Sobrido and Mostafa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures





Similar articles
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Interplay of PA-X and NS1 Proteins in Replication and Pathogenesis of a Temperature-Sensitive 2009 Pandemic H1N1 Influenza A Virus.J Virol. 2017 Aug 10;91(17):e00720-17. doi: 10.1128/JVI.00720-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637750 Free PMC article.
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza A Viruses Carrying Human or Avian Nonstructural Protein 1.J Virol. 2019 Dec 12;94(1):e00999-19. doi: 10.1128/JVI.00999-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597767 Free PMC article.
-
NS1: A Key Protein in the "Game" Between Influenza A Virus and Host in Innate Immunity.Front Cell Infect Microbiol. 2021 Jul 13;11:670177. doi: 10.3389/fcimb.2021.670177. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34327148 Free PMC article. Review.
Cited by
-
Replication kinetics, pathogenicity and virus-induced cellular responses of cattle-origin influenza A(H5N1) isolates from Texas, United States.Emerg Microbes Infect. 2025 Dec;14(1):2447614. doi: 10.1080/22221751.2024.2447614. Epub 2025 Jan 8. Emerg Microbes Infect. 2025. PMID: 39727152 Free PMC article.
-
A live attenuated NS1-deficient vaccine candidate for cattle-origin influenza A (H5N1) clade 2.3.4.4.b viruses.NPJ Vaccines. 2025 Jul 12;10(1):151. doi: 10.1038/s41541-025-01207-9. NPJ Vaccines. 2025. PMID: 40652001 Free PMC article.
-
Recombinant Influenza A Viruses Expressing Reporter Genes from the Viral NS Segment.Int J Mol Sci. 2024 Oct 1;25(19):10584. doi: 10.3390/ijms251910584. Int J Mol Sci. 2024. PMID: 39408912 Free PMC article. Review.
-
Antiviral potential of diosmin against influenza A virus.Sci Rep. 2025 May 17;15(1):17192. doi: 10.1038/s41598-025-00744-6. Sci Rep. 2025. PMID: 40382364 Free PMC article.
References
-
- Aramini J. M., Ma L. C., Zhou L., Schauder C. M., Hamilton K., Amer B. R., et al. . (2011). Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J. Biol. Chem. 286, 26050–26060. doi: 10.1074/jbc.M111.248765 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

