The SMC5/6 complex: folding chromosomes back into shape when genomes take a break
- PMID: 38375830
- PMCID: PMC10954462
- DOI: 10.1093/nar/gkae103
The SMC5/6 complex: folding chromosomes back into shape when genomes take a break
Abstract
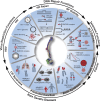
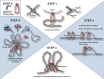
High-level folding of chromatin is a key determinant of the shape and functional state of chromosomes. During cell division, structural maintenance of chromosome (SMC) complexes such as condensin and cohesin ensure large-scale folding of chromatin into visible chromosomes. In contrast, the SMC5/6 complex plays more local and context-specific roles in the structural organization of interphase chromosomes with important implications for health and disease. Recent advances in single-molecule biophysics and cryo-electron microscopy revealed key insights into the architecture of the SMC5/6 complex and how interactions connecting the complex to chromatin components give rise to its unique repertoire of interphase functions. In this review, we provide an integrative view of the features that differentiates the SMC5/6 complex from other SMC enzymes and how these enable dramatic reorganization of DNA folding in space during DNA repair reactions and other genome transactions. Finally, we explore the mechanistic basis for the dynamic targeting of the SMC5/6 complex to damaged chromatin and its crucial role in human health.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Bell O., Tiwari V.K., Thomä N.H., Schübeler D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011; 12:554–564. - PubMed
-
- Kim E., Barth R., Dekker C. Looping the genome with SMC complexes. Annu. Rev. Biochem. 2023; 92:15–41. - PubMed
-
- Shintomi K., Inoue F., Watanabe H., Ohsumi K., Ohsugi M., Hirano T. Mitotic chromosome assembly despite nucleosome depletion in Xenopus egg extracts. Science. 2017; 356:1284–1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

