Quantitative modeling of tumor dynamics and development of drug resistance in non-small cell lung cancer patients treated with erlotinib
- PMID: 38375997
- PMCID: PMC11015077
- DOI: 10.1002/psp4.13105
Quantitative modeling of tumor dynamics and development of drug resistance in non-small cell lung cancer patients treated with erlotinib
Abstract
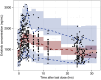
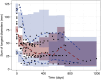
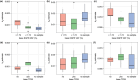
Insight into the development of treatment resistance can support the optimization of anticancer treatments. This study aims to characterize the tumor dynamics and development of drug resistance in patients with non-small cell lung cancer treated with erlotinib, and investigate the relationship between baseline circulating tumor DNA (ctDNA) data and tumor dynamics. Data obtained for the analysis included (1) intensively sampled erlotinib concentrations from 29 patients from two previous pharmacokinetic (PK) studies, and (2) tumor sizes, ctDNA measurements, and sparsely sampled erlotinib concentrations from 18 patients from the START-TKI study. A two-compartment population PK model was first developed which well-described the PK data. The PK model was subsequently applied to investigate the exposure-tumor dynamics relationship. To characterize the tumor dynamics, models accounting for intra-tumor heterogeneity and acquired resistance with or without primary resistance were investigated. Eventually, the model assumed acquired resistance only resulted in an adequate fit. Additionally, models with or without exposure-dependent treatment effect were explored, and no significant exposure-response relationship for erlotinib was identified within the observed exposure range. Subsequently, the correlation of baseline ctDNA data on EGFR and TP53 variants with tumor dynamics' parameters was explored. The analysis indicated that higher baseline plasma EGFR mutation levels correlated with increased tumor growth rates, and the inclusion of ctDNA measurements improved model fit. This result suggests that quantitative ctDNA measurements at baseline have the potential to be a predictor of anticancer treatment response. The developed model can potentially be applied to design optimal treatment regimens that better overcome resistance.
© 2024 The Authors. CPT: Pharmacometrics & Systems Pharmacology published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
The authors declared no competing interests for this work.
Figures
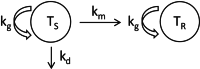
Similar articles
-
Chloroquine in combination with aptamer-modified nanocomplexes for tumor vessel normalization and efficient erlotinib/Survivin shRNA co-delivery to overcome drug resistance in EGFR-mutated non-small cell lung cancer.Acta Biomater. 2018 Aug;76:257-274. doi: 10.1016/j.actbio.2018.06.034. Epub 2018 Jun 28. Acta Biomater. 2018. PMID: 29960010
-
A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance.Cancer Med. 2017 Jan;6(1):154-162. doi: 10.1002/cam4.978. Epub 2016 Dec 20. Cancer Med. 2017. PMID: 28000387 Free PMC article.
-
Heterogeneous resistance mechanisms in an EGFR exon 19-mutated non-small cell lung cancer patient treated with erlotinib: Persistent FGFR3-mutation, localized transformation to EGFR-mutated SCLC, and acquired T790M EGFR-mutation.Lung Cancer. 2017 Nov;113:14-17. doi: 10.1016/j.lungcan.2017.08.024. Epub 2017 Sep 1. Lung Cancer. 2017. PMID: 29110841
-
Strategies to overcome acquired resistance to EGFR TKI in the treatment of non-small cell lung cancer.Clin Transl Oncol. 2019 Oct;21(10):1287-1301. doi: 10.1007/s12094-019-02075-1. Epub 2019 Mar 12. Clin Transl Oncol. 2019. PMID: 30864018 Review.
-
Characterising acquired resistance to erlotinib in non-small cell lung cancer patients.Expert Rev Respir Med. 2019 Oct;13(10):1019-1028. doi: 10.1080/17476348.2019.1656068. Epub 2019 Aug 19. Expert Rev Respir Med. 2019. PMID: 31411906 Review.
References
-
- Oliveira KCS, Ramos IB, Silva JMC, et al. Current perspectives on circulating tumor DNA, precision medicine, and personalized clinical management of cancer. Mol Cancer Res. 2020;18:517‐528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

