No significant change of N6 -methyladenosine modification landscape in mouse brain after morphine exposure
- PMID: 38376052
- PMCID: PMC10757896
- DOI: 10.1002/brb3.3350
No significant change of N6 -methyladenosine modification landscape in mouse brain after morphine exposure
Abstract
Objectives: N6 -methyladenosine (m6 A) plays a crucial role in regulating neuroplasticity and different brain functions at the posttranscriptional level. However, it remains unknown whether m6 A modification is involved in acute and chronic morphine exposure.
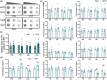
Materials and methods: In this study, we conducted a direct comparison of m6 A levels and mRNA expression of m6 A-associated factors between morphine-treated and nontreated C57BL/6 wild-type mice. We established animal models of both acute and chronic morphine treatment and confirmed the rewarding effects of chronic morphine treatment using the conditioned place preference (CPP) assay. The activation status of different brain regions in response to morphine was assessed by c-fos staining. To assess overall m6 A modification levels, we employed the m6 A dot blot assay, while mRNA levels of m6 A-associated proteins were measured using a quantitative polymerase chain reaction (qPCR) assay. These analyses were performed to investigate whether and how m6 A modification and m6 A-associated protein expression will change following morphine exposure.
Results: The overall m6 A methylation and mRNA levels of m6 A-associated proteins were not significantly altered in brain regions that were either activated or not activated during acute morphine stimulation. Similarly, the overall m6 A modification and mRNA levels of m6 A-associated proteins remained unaffected in several key brain regions associated with reward following chronic morphine exposure.
Conclusion: This study showed that the overall m6 A modification level and mRNA expression levels of m6 A-associated factors were not affected after acute and chronic morphine exposure in different brain regions, indicating m6 A modification may not be involved in brain response to morphine exposure.
Keywords: N6-methyladenosine; epigenetics; morphine; opioid; posttranscriptional RNA modification; reward.
© 2023 The Authors. Brain and Behavior published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Almeida‐Santos, A. F. , Gobira, P. H. , Souza, D. P. , Ferreira, R. C. M. , Romero, T. R. , Duarte, I. D. , Aguiar, D. C. , & Moreira, F. A. (2014). The antipsychotic aripiprazole selectively prevents the stimulant and rewarding effects of morphine in mice. European Journal of Pharmacology, 742, 139–144. 10.1016/j.ejphar.2014.09.004 - DOI - PubMed
-
- Bazov, I. , Sarkisyan, D. , Kononenko, O. , Watanabe, H. , Taqi, M. M. , Stålhandske, L. , Verbeek, D. S. , Mulder, J. , Rajkowska, G. , Sheedy, D. , Kril, J. , Sun, X. , Syvänen, A.‐C. , Yakovleva, T. , & Bakalkin, G. (2018). Neuronal expression of opioid gene is controlled by dual epigenetic and transcriptional mechanism in human brain. Cerebral Cortex, 28(9), 3129–3142. 10.1093/cercor/bhx181 - DOI - PMC - PubMed
-
- Bokar, J. A. , Shambaugh, M. E. , Polayes, D. , Matera, A. G. , & Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet‐binding subunit of the human mRNA (N6‐adenosine)‐methyltransferase. RNA, 3(11), 1233–1247. https://www.ncbi.nlm.nih.gov/pubmed/9409616 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 31922026/National Natural Science Foundation of China
- U20A2016/National Natural Science Foundation of China
- 2018YFA0801401/National Key Research and Development Program of China
- ZDSYS20190902093601675/Shenzhen Science and Technology Innovation Program
- 2019B151502007/Basic and Applied Basic Research Foundation of Guangdong Province
LinkOut - more resources
Full Text Sources

