Benchmarking informatics approaches for virus discovery: caution is needed when combining in silico identification methods
- PMID: 38376167
- PMCID: PMC10949488
- DOI: 10.1128/msystems.01105-23
Benchmarking informatics approaches for virus discovery: caution is needed when combining in silico identification methods
Abstract
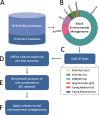
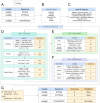
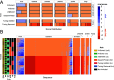
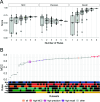
Understanding the ecological impacts of viruses on natural and engineered ecosystems relies on the accurate identification of viral sequences from community sequencing data. To maximize viral recovery from metagenomes, researchers frequently combine viral identification tools. However, the effectiveness of this strategy is unknown. Here, we benchmarked combinations of six widely used informatics tools for viral identification and analysis (VirSorter, VirSorter2, VIBRANT, DeepVirFinder, CheckV, and Kaiju), called "rulesets." Rulesets were tested against mock metagenomes composed of taxonomically diverse sequence types and diverse aquatic metagenomes to assess the effects of the degree of viral enrichment and habitat on tool performance. We found that six rulesets achieved equivalent accuracy [Matthews Correlation Coefficient (MCC) = 0.77, Padj ≥ 0.05]. Each contained VirSorter2, and five used our "tuning removal" rule designed to remove non-viral contamination. While DeepVirFinder, VIBRANT, and VirSorter were each found once in these high-accuracy rulesets, they were not found in combination with each other: combining tools does not lead to optimal performance. Our validation suggests that the MCC plateau at 0.77 is partly caused by inaccurate labeling within reference sequence databases. In aquatic metagenomes, our highest MCC ruleset identified more viral sequences in virus-enriched (44%-46%) than in cellular metagenomes (7%-19%). While improved algorithms may lead to more accurate viral identification tools, this should be done in tandem with careful curation of sequence databases. We recommend using the VirSorter2 ruleset and our empirically derived tuning removal rule. Our analysis provides insight into methods for in silico viral identification and will enable more robust viral identification from metagenomic data sets.
Importance: The identification of viruses from environmental metagenomes using informatics tools has offered critical insights in microbial ecology. However, it remains difficult for researchers to know which tools optimize viral recovery for their specific study. In an attempt to recover more viruses, studies are increasingly combining the outputs from multiple tools without validating this approach. After benchmarking combinations of six viral identification tools against mock metagenomes and environmental samples, we found that these tools should only be combined cautiously. Two to four tool combinations maximized viral recovery and minimized non-viral contamination compared with either the single-tool or the five- to six-tool ones. By providing a rigorous overview of the behavior of in silico viral identification strategies and a pipeline to replicate our process, our findings guide the use of existing viral identification tools and offer a blueprint for feature engineering of new tools that will lead to higher-confidence viral discovery in microbiome studies.
Keywords: bacteriophages; metagenomics; microbial ecology; viral discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. BioScience 49:781–788. 10.2307/1313569. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

