Inter-species interactions between two bacterial flower commensals and a floral pathogen reduce disease incidence and alter pathogen activity
- PMID: 38376185
- PMCID: PMC10936193
- DOI: 10.1128/mbio.00213-24
Inter-species interactions between two bacterial flower commensals and a floral pathogen reduce disease incidence and alter pathogen activity
Abstract
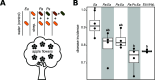
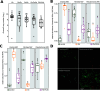
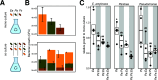
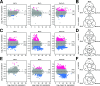
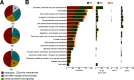
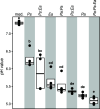
Flowers are colonized by a diverse community of microorganisms that can alter plant health and interact with floral pathogens. Erwinia amylovora is a flower-inhabiting bacterium and a pathogen that infects different plant species, including Malus × domestica (apple). Previously, we showed that the co-inoculation of two bacterial strains, members of the genera Pseudomonas and Pantoea, isolated from apple flowers, reduced disease incidence caused by this floral pathogen. Here, we decipher the ecological interactions between the two flower-associated bacteria and E. amylovora in field experimentation and in vitro co-cultures. The two flower commensal strains did not competitively exclude E. amylovora from the stigma habitat, as both bacteria and the pathogen co-existed on the stigma of apple flowers and in vitro. This suggests that plant protection might be mediated by other mechanisms than competitive niche exclusion. Using a synthetic stigma exudation medium, ternary co-culture of the bacterial strains led to a substantial alteration of gene expression in both the pathogen and the two microbiota members. Importantly, the gene expression profiles for the ternary co-culture were not just additive from binary co-cultures, suggesting that some functions only emerged in multipartite co-culture. Additionally, the ternary co-culture of the strains resulted in a stronger acidification of the growth milieu than mono- or binary co-cultures, pointing to another emergent property of co-inoculation. Our study emphasizes the critical role of emergent properties mediated by inter-species interactions within the plant holobiont and their potential impact on plant health and pathogen behavior.
Importance: Fire blight, caused by Erwinia amylovora, is one of the most important plant diseases of pome fruits. Previous work largely suggested plant microbiota commensals suppressed disease by antagonizing pathogen growth. However, inter-species interactions of multiple flower commensals and their influence on pathogen activity and behavior have not been well studied. Here, we show that co-inoculating two bacterial strains that naturally colonize the apple flowers reduces disease incidence. We further demonstrate that the interactions between these two microbiota commensals and the floral pathogen led to the emergence of new gene expression patterns and a strong alteration of the external pH, factors that may modify the pathogen's behavior. Our findings emphasize the critical role of emergent properties mediated by inter-species interactions between plant microbiota and plant pathogens and their impact on plant health.
Keywords: Erwinia amylovora; Malus domestica; Pantoea; Pseudomonas; co-culture; co-inoculation; emergent property; fire blight; meta-transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Vannette RL. 2020. The floral microbiome: plant, pollinator, and microbial perspectives. Annu Rev Ecol Evol Syst 51:363–386. doi:10.1146/annurev-ecolsys-011720-013401 - DOI
-
- Steven B, Huntley RB, Zeng Q. 2018. The influence of flower anatomy and apple cultivar on the apple flower phytobiome. Phytobiomes J 2:171–179. doi:10.1094/PBIOMES-03-18-0015-R - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
