Ubiquitin-specific proteinase 1 stabilizes PRRSV nonstructural protein Nsp1β to promote viral replication by regulating K48 ubiquitination
- PMID: 38376196
- PMCID: PMC10949481
- DOI: 10.1128/jvi.01686-23
Ubiquitin-specific proteinase 1 stabilizes PRRSV nonstructural protein Nsp1β to promote viral replication by regulating K48 ubiquitination
Abstract
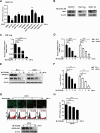
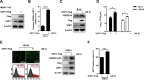
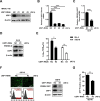
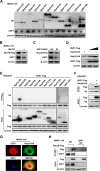
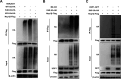
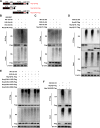
The porcine reproductive and respiratory syndrome virus (PRRSV) can lead to severe reproductive problems in sows, pneumonia in weaned piglets, and increased mortality, significantly negatively impacting the economy. Post-translational changes are essential for the host-dependent replication and long-term infection of PRRSV. Uncertainty surrounds the function of the ubiquitin network in PRRSV infection. Here, we screened 10 deubiquitinating enzyme inhibitors and found that the ubiquitin-specific proteinase 1 (USP1) inhibitor ML323 significantly inhibited PRRSV replication in vitro. Importantly, we found that USP1 interacts with nonstructural protein 1β (Nsp1β) and deubiquitinates its K48 to increase protein stability, thereby improving PRRSV replication and viral titer. Among them, lysine at position 45 is essential for Nsp1β protein stability. In addition, deficiency of USP1 significantly reduced viral replication. Moreover, ML323 loses antagonism to PRRSV rSD16-K45R. This study reveals the mechanism by which PRRSV recruits the host factor USP1 to promote viral replication, providing a new target for PRRSV defense.IMPORTANCEDeubiquitinating enzymes are critical factors in regulating host innate immunity. The porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural protein 1β (Nsp1β) is essential for producing viral subgenomic mRNA and controlling the host immune system. The host inhibits PRRSV proliferation by ubiquitinating Nsp1β, and conversely, PRRSV recruits the host protein ubiquitin-specific proteinase 1 (USP1) to remove this restriction. Our results demonstrate the binding of USP1 to Nsp1β, revealing a balance of antagonism between PRRSV and the host. Our research identifies a brand-new PRRSV escape mechanism from the immune response.
Keywords: Nsp1β; PRRSV; USP1; deubiquitination; immune escape.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
THAP11-mediated K48- and K63-linked ubiquitination is essential for the degradation of porcine reproductive and respiratory syndrome virus nonstructural protein 1β.Cell Mol Life Sci. 2025 Jun 23;82(1):246. doi: 10.1007/s00018-025-05760-3. Cell Mol Life Sci. 2025. PMID: 40548980 Free PMC article.
-
Porcine Reproductive and Respiratory Syndrome Virus nsp1β Stabilizes HIF-1α to Enhance Viral Replication.Microbiol Spectr. 2022 Dec 21;10(6):e0317322. doi: 10.1128/spectrum.03173-22. Epub 2022 Nov 23. Microbiol Spectr. 2022. PMID: 36416550 Free PMC article.
-
Porcine Reproductive and Respiratory Syndrome Virus Nonstructural Protein 1 Beta Interacts with Nucleoporin 62 To Promote Viral Replication and Immune Evasion.J Virol. 2019 Jun 28;93(14):e00469-19. doi: 10.1128/JVI.00469-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31043527 Free PMC article.
-
N-Acetyltransferase 9 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Proliferation by N-Terminal Acetylation of the Structural Protein GP5.Microbiol Spectr. 2023 Feb 14;11(1):e0244222. doi: 10.1128/spectrum.02442-22. Epub 2023 Jan 25. Microbiol Spectr. 2023. PMID: 36695606 Free PMC article.
-
Discovery of ML323 as a Novel Inhibitor of the USP1/UAF1 Deubiquitinase Complex.2012 Oct 23 [updated 2014 Sep 18]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. 2012 Oct 23 [updated 2014 Sep 18]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. PMID: 25506968 Free Books & Documents. Review.
Cited by
-
THAP11-mediated K48- and K63-linked ubiquitination is essential for the degradation of porcine reproductive and respiratory syndrome virus nonstructural protein 1β.Cell Mol Life Sci. 2025 Jun 23;82(1):246. doi: 10.1007/s00018-025-05760-3. Cell Mol Life Sci. 2025. PMID: 40548980 Free PMC article.
-
Hyperoside inhibits PRRSV proliferation via the TLR4/NF-κB and p62-Nrf2-Keap1 signaling pathways, mediating inflammation and autophagy.Microbiol Spectr. 2025 Aug 5;13(8):e0310724. doi: 10.1128/spectrum.03107-24. Epub 2025 Jun 12. Microbiol Spectr. 2025. PMID: 40503831 Free PMC article.
-
Identification and biophysical characterization of epitope atlas of Porcine Reproductive and Respiratory Syndrome Virus.Comput Struct Biotechnol J. 2024 Sep 10;23:3348-3357. doi: 10.1016/j.csbj.2024.08.029. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39310279 Free PMC article.
-
Effects of the commensal microbiota on spleen and mesenteric lymph node immune function: investigation in a germ-free piglet model.Front Microbiol. 2024 Jun 12;15:1398631. doi: 10.3389/fmicb.2024.1398631. eCollection 2024. Front Microbiol. 2024. PMID: 38933022 Free PMC article.
-
Strategies and scheming: the war between PRRSV and host cells.Virol J. 2025 Jun 11;22(1):191. doi: 10.1186/s12985-025-02685-y. Virol J. 2025. PMID: 40500743 Free PMC article. Review.
References
-
- Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi:10.2460/javma.2005.227.385 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

