Nanopore adaptive sampling effectively enriches bacterial plasmids
- PMID: 38376263
- PMCID: PMC10949517
- DOI: 10.1128/msystems.00945-23
Nanopore adaptive sampling effectively enriches bacterial plasmids
Abstract
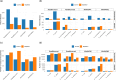
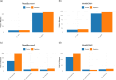
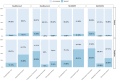
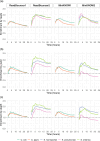
Bacterial plasmids play a major role in the spread of antibiotic resistance genes. However, their characterization via DNA sequencing suffers from the low abundance of plasmid DNA in those samples. Although sample preparation methods can enrich the proportion of plasmid DNA before sequencing, these methods are expensive and laborious, and they might introduce a bias by enriching only for specific plasmid DNA sequences. Nanopore adaptive sampling could overcome these issues by rejecting uninteresting DNA molecules during the sequencing process. In this study, we assess the application of adaptive sampling for the enrichment of low-abundant plasmids in known bacterial isolates using two different adaptive sampling tools. We show that a significant enrichment can be achieved even on expired flow cells. By applying adaptive sampling, we also improve the quality of de novo plasmid assemblies and reduce the sequencing time. However, our experiments also highlight issues with adaptive sampling if target and non-target sequences span similar regions.
Importance: Antimicrobial resistance causes millions of deaths every year. Mobile genetic elements like bacterial plasmids are key drivers for the dissemination of antimicrobial resistance genes. This makes the characterization of plasmids via DNA sequencing an important tool for clinical microbiologists. Since plasmids are often underrepresented in bacterial samples, plasmid sequencing can be challenging and laborious. To accelerate the sequencing process, we evaluate nanopore adaptive sampling as an in silico method for the enrichment of low-abundant plasmids. Our results show the potential of this cost-efficient method for future plasmid research but also indicate issues that arise from using reference sequences.
Keywords: adaptive sampling; bacteria; enrichment; nanopore sequencing; plasmid; readuntil.
Conflict of interest statement
J.U.U. and B.Y.R. have filed a patent application on selective nanopore sequencing approaches.
Figures

Similar articles
-
Impact of microbiological molecular methodologies on adaptive sampling using nanopore sequencing in metagenomic studies.Environ Microbiome. 2025 May 5;20(1):47. doi: 10.1186/s40793-025-00704-7. Environ Microbiome. 2025. PMID: 40325409 Free PMC article.
-
Tracking of Antibiotic Resistance Transfer and Rapid Plasmid Evolution in a Hospital Setting by Nanopore Sequencing.mSphere. 2020 Aug 19;5(4):e00525-20. doi: 10.1128/mSphere.00525-20. mSphere. 2020. PMID: 32817379 Free PMC article.
-
Recovery of small plasmid sequences via Oxford Nanopore sequencing.Microb Genom. 2021 Aug;7(8):000631. doi: 10.1099/mgen.0.000631. Microb Genom. 2021. PMID: 34431763 Free PMC article.
-
Direct sequencing of insect symbionts via nanopore adaptive sampling.Curr Opin Insect Sci. 2024 Feb;61:101135. doi: 10.1016/j.cois.2023.101135. Epub 2023 Nov 4. Curr Opin Insect Sci. 2024. PMID: 37926187 Free PMC article. Review.
-
Global epistasis in plasmid-mediated antimicrobial resistance.Mol Syst Biol. 2024 Apr;20(4):311-320. doi: 10.1038/s44320-024-00012-1. Epub 2024 Feb 26. Mol Syst Biol. 2024. PMID: 38409539 Free PMC article. Review.
Cited by
-
ReadCurrent: a VDCNN-based tool for fast and accurate nanopore selective sequencing.Brief Bioinform. 2024 Jul 25;25(5):bbae435. doi: 10.1093/bib/bbae435. Brief Bioinform. 2024. PMID: 39226890 Free PMC article.
-
Improving Nanopore sequencing-based core genome MLST for global infection control: a strategy for GC-rich pathogens like Burkholderia pseudomallei.J Clin Microbiol. 2025 Mar 12;63(3):e0156924. doi: 10.1128/jcm.01569-24. Epub 2025 Feb 6. J Clin Microbiol. 2025. PMID: 39912668 Free PMC article.
-
Plasmidome of Salmonella enterica serovar Infantis recovered from surface waters in a major agricultural region for leafy greens in California.PLoS One. 2024 Dec 30;19(12):e0316466. doi: 10.1371/journal.pone.0316466. eCollection 2024. PLoS One. 2024. PMID: 39775564 Free PMC article.
-
Impact of microbiological molecular methodologies on adaptive sampling using nanopore sequencing in metagenomic studies.Environ Microbiome. 2025 May 5;20(1):47. doi: 10.1186/s40793-025-00704-7. Environ Microbiome. 2025. PMID: 40325409 Free PMC article.
-
Targeted sequencing of Enterobacterales bacteria using CRISPR-Cas9 enrichment and Oxford Nanopore Technologies.mSystems. 2025 Feb 18;10(2):e0141324. doi: 10.1128/msystems.01413-24. Epub 2025 Jan 8. mSystems. 2025. PMID: 39772804 Free PMC article.
References
-
- O’Neil J. 2014. Tackling a crisis for the health and wealth of nations. World Health Organization.
-
- O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

