Genomic insights into virulence, antimicrobial resistance, and adaptation acumen of Escherichia coli isolated from an urban environment
- PMID: 38376265
- PMCID: PMC10936179
- DOI: 10.1128/mbio.03545-23
Genomic insights into virulence, antimicrobial resistance, and adaptation acumen of Escherichia coli isolated from an urban environment
Abstract
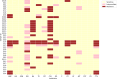
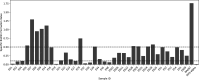
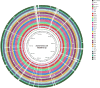
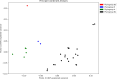
Populations of common commensal bacteria such as Escherichia coli undergo genetic changes by the acquisition of certain virulence and antimicrobial resistance (AMR) encoding genetic elements leading to the emergence of pathogenic strains capable of surviving in the previously uninhabited or protected niches. These bacteria are also reported to be prevalent in the environment where they survive by adopting various recombination strategies to counter microflora of the soil and water, under constant selection pressure(s). In this study, we performed molecular characterization, phenotypic AMR analysis, and whole genome sequencing (WGS) of E. coli (n = 37) isolated from soil and surface water representing the urban and peri-urban areas. The primary aim of this study was to understand the genetic architecture and pathogenic acumen exhibited by environmental E. coli. WGS-based analysis entailing resistome and virulome profiling indicated the presence of various virulence (adherence, iron uptake, and toxins) and AMR encoding genes, including blaNDM-5 in the environmental isolates. A majority of our isolates belonged to phylogroup B1 (73%). A few isolates in our collection were of sequence type(s) (ST) 58 and 224 that could have emerged recently as clonal lineages and might pose risk of infection/transmission. Mobile genetic elements (MGEs) such as plasmids (predominantly) of the IncF family, prophages, pipolins, and insertion elements such as IS1 and IS5 were also observed to exist, which may presumably aid in the propagation of genes encoding resistance against antimicrobial drugs. The observed high prevalence of MGEs associated with multidrug resistance in pathogenic E. coli isolates belonging to the phylogroup B1 underscores the need for extended surveillance to keep track of and prevent the transmission of the bacterium to certain vulnerable human and animal populations.
Importance: Evolutionary patterns of E. coli bacteria convey that they evolve into highly pathogenic forms by acquiring fitness advantages, such as AMR, and various virulence factors through the horizontal gene transfer (HGT)-mediated acquisition of MGEs. However, limited research on the genetic profiles of environmental E. coli, particularly from India, hinders our understanding of their transition to pathogenic forms and impedes the adoption of a comprehensive approach to address the connection between environmentally dwelling E. coli populations and human and veterinary public health. This study focuses on high-resolution genomic analysis of the environmental E. coli isolates aiming to understand the genetic similarities and differences among isolates from different environmental niches and uncover the survival strategies employed by these bacteria to thrive in their surroundings. Our approach involved molecular characterization of environmental samples using PCR-based DNA fingerprinting and subsequent WGS analysis. This multidisciplinary approach is likely to provide valuable insights into the understanding of any potential spill-over to human and animal populations and locales. Investigating these environmental isolates has significant potential for developing epidemiological strategies against transmission and understanding niche-specific evolutionary patterns.
Keywords: Escherichia coli; antimicrobial resistance; environment; genome analysis; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Whole genome analysis of multidrug-resistant Escherichia coli isolate collected from drinking water in Armenia revealed the plasmid-borne mcr-1.1-mediated colistin resistance.Microbiol Spectr. 2024 Oct 3;12(10):e0075124. doi: 10.1128/spectrum.00751-24. Epub 2024 Aug 21. Microbiol Spectr. 2024. PMID: 39166856 Free PMC article.
-
Genome Informatics and Machine Learning-Based Identification of Antimicrobial Resistance-Encoding Features and Virulence Attributes in Escherichia coli Genomes Representing Globally Prevalent Lineages, Including High-Risk Clonal Complexes.mBio. 2021 Feb 22;13(1):e0379621. doi: 10.1128/mbio.03796-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164570 Free PMC article.
-
Antibiotic resistance genes, mobile elements, virulence genes, and phages in cultivated ESBL-producing Escherichia coli of poultry origin in Kwara State, North Central Nigeria.Int J Food Microbiol. 2023 Mar 16;389:110086. doi: 10.1016/j.ijfoodmicro.2023.110086. Epub 2023 Jan 21. Int J Food Microbiol. 2023. PMID: 36738714
-
Escherichia coli: An arduous voyage from commensal to Antibiotic-resistance.Microb Pathog. 2025 Jan;198:107173. doi: 10.1016/j.micpath.2024.107173. Epub 2024 Nov 27. Microb Pathog. 2025. PMID: 39608506 Review.
-
Drivers of virulence and antimicrobial resistance in Gram-negative bacteria in different settings: A genomic perspective.Infect Genet Evol. 2024 Oct;124:105666. doi: 10.1016/j.meegid.2024.105666. Epub 2024 Sep 4. Infect Genet Evol. 2024. PMID: 39242067 Review.
Cited by
-
Emerging multi-drug resistant and extended-spectrum β-lactamase (ESBL)-positive enterotoxigenic E. coli (ETEC) clones circulating in aquatic environments and in patients.One Health. 2025 Jan 10;20:100968. doi: 10.1016/j.onehlt.2025.100968. eCollection 2025 Jun. One Health. 2025. PMID: 39898314 Free PMC article.
-
Whole genome analysis of multidrug-resistant Escherichia coli isolate collected from drinking water in Armenia revealed the plasmid-borne mcr-1.1-mediated colistin resistance.Microbiol Spectr. 2024 Oct 3;12(10):e0075124. doi: 10.1128/spectrum.00751-24. Epub 2024 Aug 21. Microbiol Spectr. 2024. PMID: 39166856 Free PMC article.
-
Antimicrobial Resistant Staphylococcus spp., Escherichia coli, and Salmonella spp. in Food Handlers: A Global Review of Persistence, Transmission, and Mitigation Challenges.Pathogens. 2025 May 18;14(5):496. doi: 10.3390/pathogens14050496. Pathogens. 2025. PMID: 40430816 Free PMC article. Review.
-
The relationship between integrons, antibiotic resistance genes and SXT resistance in Shigella flexneri strains.Am J Transl Res. 2024 May 15;16(5):1925-1934. doi: 10.62347/SNRQ6766. eCollection 2024. Am J Transl Res. 2024. PMID: 38883348 Free PMC article.
-
The Trade-Off Between Sanitizer Resistance and Virulence Genes: Genomic Insights into E. coli Adaptation.Antibiotics (Basel). 2025 Mar 11;14(3):291. doi: 10.3390/antibiotics14030291. Antibiotics (Basel). 2025. PMID: 40149102 Free PMC article.
References
-
- Thanh Duy P, Thi Nguyen TN, Vu Thuy D, Chung The H, Alcock F, Boinett C, Dan Thanh HN, Thanh Tuyen H, Thwaites GE, Rabaa MA, Baker S. 2020. Commensal Escherichia coli are a reservoir for the transfer of XDR plasmids into epidemic fluoroquinolone-resistant Shigella sonnei. Nat Microbiol 5:256–264. doi:10.1038/s41564-019-0645-9 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

