The HIV-1 reservoir landscape in persistent elite controllers and transient elite controllers
- PMID: 38376918
- PMCID: PMC11014653
- DOI: 10.1172/JCI174215
The HIV-1 reservoir landscape in persistent elite controllers and transient elite controllers
Abstract
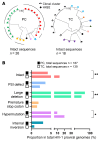
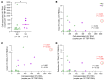
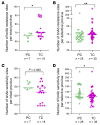
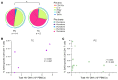
BACKGROUNDPersistent controllers (PCs) maintain antiretroviral-free HIV-1 control indefinitely over time, while transient controllers (TCs) eventually lose virological control. It is essential to characterize the quality of the HIV reservoir in terms of these phenotypes in order to identify the factors that lead to HIV progression and to open new avenues toward an HIV cure.METHODSThe characterization of HIV-1 reservoir from peripheral blood mononuclear cells was performed using next-generation sequencing techniques, such as full-length individual and matched integration site proviral sequencing (FLIP-Seq; MIP-Seq).RESULTSPCs and TCs, before losing virological control, presented significantly lower total, intact, and defective proviruses compared with those of participants on antiretroviral therapy (ART). No differences were found in total and defective proviruses between PCs and TCs. However, intact provirus levels were lower in PCs compared with TCs; indeed the intact/defective HIV-DNA ratio was significantly higher in TCs. Clonally expanded intact proviruses were found only in PCs and located in centromeric satellite DNA or zinc-finger genes, both associated with heterochromatin features. In contrast, sampled intact proviruses were located in permissive genic euchromatic positions in TCs.CONCLUSIONSThese results suggest the need for, and can give guidance to, the design of future research to identify a distinct proviral landscape that may be associated with the persistent control of HIV-1 without ART.FUNDINGInstituto de Salud Carlos III (FI17/00186, FI19/00083, MV20/00057, PI18/01532, PI19/01127 and PI22/01796), Gilead Fellowships (GLD22/00147). NIH grants AI155171, AI116228, AI078799, HL134539, DA047034, MH134823, amfAR ARCHE and the Bill and Melinda Gates Foundation.
Keywords: AIDS vaccine; AIDS/HIV; Virology.
Conflict of interest statement
Figures






References
MeSH terms
Substances
Grants and funding
- R01 AI078799/AI/NIAID NIH HHS/United States
- K24 AI155233/AI/NIAID NIH HHS/United States
- R01 HL134539/HL/NHLBI NIH HHS/United States
- UM1 AI164570/AI/NIAID NIH HHS/United States
- R61 DA047034/DA/NIDA NIH HHS/United States
- UM1 AI164562/AI/NIAID NIH HHS/United States
- U01 AI135940/AI/NIAID NIH HHS/United States
- R37 AI155171/AI/NIAID NIH HHS/United States
- R33 DA047034/DA/NIDA NIH HHS/United States
- R01 AI152979/AI/NIAID NIH HHS/United States
- R01 MH134823/MH/NIMH NIH HHS/United States
- R01 AI176579/AI/NIAID NIH HHS/United States
- R01 AI130005/AI/NIAID NIH HHS/United States
- U01 AI117841/AI/NIAID NIH HHS/United States
- R01 DK120387/DK/NIDDK NIH HHS/United States
- UM1 AI164560/AI/NIAID NIH HHS/United States
- R33 AI116228/AI/NIAID NIH HHS/United States
- R21 AI116228/AI/NIAID NIH HHS/United States
- UM1 AI164566/AI/NIAID NIH HHS/United States

