Mitochondrial and Cytosolic One-Carbon Metabolism Is a Targetable Metabolic Vulnerability in Cisplatin-Resistant Ovarian Cancer
- PMID: 38377173
- PMCID: PMC11150100
- DOI: 10.1158/1535-7163.MCT-23-0550
Mitochondrial and Cytosolic One-Carbon Metabolism Is a Targetable Metabolic Vulnerability in Cisplatin-Resistant Ovarian Cancer
Abstract
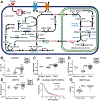
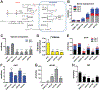
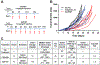
One-carbon (C1) metabolism is compartmentalized between the cytosol and mitochondria with the mitochondrial C1 pathway as the major source of glycine and C1 units for cellular biosynthesis. Expression of mitochondrial C1 genes including SLC25A32, serine hydroxymethyl transferase (SHMT) 2, 5,10-methylene tetrahydrofolate dehydrogenase 2, and 5,10-methylene tetrahydrofolate dehydrogenase 1-like was significantly elevated in primary epithelial ovarian cancer (EOC) specimens compared with normal ovaries. 5-Substituted pyrrolo[3,2-d]pyrimidine antifolates (AGF347, AGF359, AGF362) inhibited proliferation of cisplatin-sensitive (A2780, CaOV3, IGROV1) and cisplatin-resistant (A2780-E80, SKOV3) EOC cells. In SKOV3 and A2780-E80 cells, colony formation was inhibited. AGF347 induced apoptosis in SKOV3 cells. In IGROV1 cells, AGF347 was transported by folate receptor (FR) α. AGF347 was also transported into IGROV1 and SKOV3 cells by the proton-coupled folate transporter (SLC46A1) and the reduced folate carrier (SLC19A1). AGF347 accumulated to high levels in the cytosol and mitochondria of SKOV3 cells. By targeted metabolomics with [2,3,3-2H]L-serine, AGF347, AGF359, and AGF362 inhibited SHMT2 in the mitochondria. In the cytosol, SHMT1 and de novo purine biosynthesis (i.e., glycinamide ribonucleotide formyltransferase, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase) were targeted; AGF359 also inhibited thymidylate synthase. Antifolate treatments of SKOV3 cells depleted cellular glycine, mitochondrial NADH and glutathione, and showed synergistic in vitro inhibition toward SKOV3 and A2780-E80 cells when combined with cisplatin. In vivo studies with subcutaneous SKOV3 EOC xenografts in SCID mice confirmed significant antitumor efficacy of AGF347. Collectively, our studies demonstrate a unique metabolic vulnerability in EOC involving mitochondrial and cytosolic C1 metabolism, which offers a promising new platform for therapy.
©2024 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures


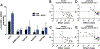
References
-
- Stover PJ. Physiology of folate and vitamin B12 in health and disease. Nutrition reviews 2004;62(6 Pt 2):S3–12; discussion S3. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

