The global RNA-RNA interactome of Klebsiella pneumoniae unveils a small RNA regulator of cell division
- PMID: 38377209
- PMCID: PMC10907235
- DOI: 10.1073/pnas.2317322121
The global RNA-RNA interactome of Klebsiella pneumoniae unveils a small RNA regulator of cell division
Abstract
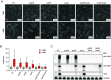
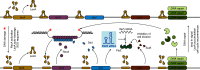
The ubiquitous RNA chaperone Hfq is involved in the regulation of key biological processes in many species across the bacterial kingdom. In the opportunistic human pathogen Klebsiella pneumoniae, deletion of the hfq gene affects the global transcriptome, virulence, and stress resistance; however, the ligands of the major RNA-binding protein in this species have remained elusive. In this study, we have combined transcriptomic, co-immunoprecipitation, and global RNA interactome analyses to compile an inventory of conserved and species-specific RNAs bound by Hfq and to monitor Hfq-mediated RNA-RNA interactions. In addition to dozens of RNA-RNA pairs, our study revealed an Hfq-dependent small regulatory RNA (sRNA), DinR, which is processed from the 3' terminal portion of dinI mRNA. Transcription of dinI is controlled by the master regulator of the SOS response, LexA. As DinR accumulates in K. pneumoniae in response to DNA damage, the sRNA represses translation of the ftsZ transcript by occupation of the ribosome binding site. Ectopic overexpression of DinR causes depletion of ftsZ mRNA and inhibition of cell division, while deletion of dinR antagonizes cell elongation in the presence of DNA damage. Collectively, our work highlights the important role of RNA-based gene regulation in K. pneumoniae and uncovers the central role of DinR in LexA-controlled division inhibition during the SOS response.
Keywords: Hfq; Klebsiella pneumoniae; RIL-seq; SOS response; small RNA.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





Similar articles
-
RNA interactome of hypervirulent Klebsiella pneumoniae reveals a small RNA inhibitor of capsular mucoviscosity and virulence.Nat Commun. 2024 Aug 13;15(1):6946. doi: 10.1038/s41467-024-51213-z. Nat Commun. 2024. PMID: 39138169 Free PMC article.
-
Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells.Elife. 2021 Feb 22;10:e64207. doi: 10.7554/eLife.64207. Elife. 2021. PMID: 33616037 Free PMC article.
-
Identification of BvgA-Dependent and BvgA-Independent Small RNAs (sRNAs) in Bordetella pertussis Using the Prokaryotic sRNA Prediction Toolkit ANNOgesic.Microbiol Spectr. 2021 Oct 31;9(2):e0004421. doi: 10.1128/Spectrum.00044-21. Epub 2021 Sep 22. Microbiol Spectr. 2021. PMID: 34550019 Free PMC article.
-
Cycling of RNAs on Hfq.RNA Biol. 2013 Apr;10(4):619-26. doi: 10.4161/rna.24044. Epub 2013 Mar 6. RNA Biol. 2013. PMID: 23466677 Free PMC article. Review.
-
Hfq chaperone brings speed dating to bacterial sRNA.Wiley Interdiscip Rev RNA. 2018 Jul;9(4):e1475. doi: 10.1002/wrna.1475. Epub 2018 Apr 6. Wiley Interdiscip Rev RNA. 2018. PMID: 29633565 Free PMC article. Review.
Cited by
-
An sRNA overexpression library reveals AbnZ as a negative regulator of an essential translocation module in Caulobacter crescentus.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1139. doi: 10.1093/nar/gkae1139. Nucleic Acids Res. 2025. PMID: 39657128 Free PMC article.
-
RNA interactome of hypervirulent Klebsiella pneumoniae reveals a small RNA inhibitor of capsular mucoviscosity and virulence.Nat Commun. 2024 Aug 13;15(1):6946. doi: 10.1038/s41467-024-51213-z. Nat Commun. 2024. PMID: 39138169 Free PMC article.
-
Biological Insights from RNA-RNA Interactomes in Bacteria, as Revealed by RIL-seq.Methods Mol Biol. 2025;2866:189-206. doi: 10.1007/978-1-0716-4192-7_11. Methods Mol Biol. 2025. PMID: 39546204 Review.
-
Image-based quantification of Candida albicans filamentation and hyphal length using the open-source visual programming language JIPipe.FEMS Yeast Res. 2025 Jan 30;25:foaf011. doi: 10.1093/femsyr/foaf011. FEMS Yeast Res. 2025. PMID: 40082735 Free PMC article.
-
RIL-seq reveals extensive involvement of small RNAs in virulence and capsule regulation in hypervirulent Klebsiella pneumoniae.Nucleic Acids Res. 2024 Aug 27;52(15):9119-9138. doi: 10.1093/nar/gkae440. Nucleic Acids Res. 2024. PMID: 38804271 Free PMC article.
References
-
- Tacconelli E., et al. , Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). - PubMed
-
- Sommer M. O. A., Munck C., Toft-Kehler R. V., Andersson D. I., Prediction of antibiotic resistance: Time for a new preclinical paradigm? Nat. Rev. Microbiol. 15, 689–696 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

