Metagenomic assembled genomes indicated the potential application of hypersaline microbiome for plant growth promotion and stress alleviation in salinized soils
- PMID: 38377278
- PMCID: PMC10949518
- DOI: 10.1128/msystems.01050-23
Metagenomic assembled genomes indicated the potential application of hypersaline microbiome for plant growth promotion and stress alleviation in salinized soils
Abstract
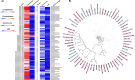
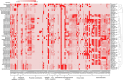
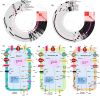
Climate change is causing unpredictable seasonal variations globally. Due to the continuously increasing earth's surface temperature, the rate of water evaporation is enhanced, conceiving a problem of soil salinization, especially in arid and semi-arid regions. The accumulation of salt degrades soil quality, impairs plant growth, and reduces agricultural yields. Salt-tolerant, plant-growth-promoting microorganisms may offer a solution, enhancing crop productivity and soil fertility in salinized areas. In the current study, genome-resolved metagenomic analysis has been performed to investigate the salt-tolerating and plant growth-promoting potential of two hypersaline ecosystems, Sambhar Lake and Drang Mine. The samples were co-assembled independently by Megahit, MetaSpades, and IDBA-UD tools. A total of 67 metagenomic assembled genomes (MAGs) were reconstructed following the binning process, including 15 from Megahit, 26 from MetaSpades, and 26 from IDBA_UD assembly tools. As compared to other assemblers, the MAGs obtained by MetaSpades were of superior quality, with a completeness range of 12.95%-96.56% and a contamination range of 0%-8.65%. The medium and high-quality MAGs from MetaSpades, upon functional annotation, revealed properties such as salt tolerance (91.3%), heavy metal tolerance (95.6%), exopolysaccharide (95.6%), and antioxidant (60.86%) biosynthesis. Several plant growth-promoting attributes, including phosphate solubilization and indole-3-acetic acid (IAA) production, were consistently identified across all obtained MAGs. Conversely, characteristics such as iron acquisition and potassium solubilization were observed in a substantial majority, specifically 91.3%, of the MAGs. The present study indicates that hypersaline microflora can be used as bio-fertilizing agents for agricultural practices in salinized areas by alleviating prevalent stresses.
Importance: The strategic implementation of metagenomic assembled genomes (MAGs) in exploring the properties and harnessing microorganisms from ecosystems like hypersaline niches has transformative potential in agriculture. This approach promises to redefine our comprehension of microbial diversity and its ecosystem roles. Recovery and decoding of MAGs unlock genetic resources, enabling the development of new solutions for agricultural challenges. Enhanced understanding of these microbial communities can lead to more efficient nutrient cycling, pest control, and soil health maintenance. Consequently, traditional agricultural practices can be improved, resulting in increased yields, reduced environmental impacts, and heightened sustainability. MAGs offer a promising avenue for sustainable agriculture, bridging the gap between cutting-edge genomics and practical field applications.
Keywords: heavy metal stress; hypersaline ecosystems; metagenomic assembled genomes; plant growth promotion; salinized soil; salt stress.
Conflict of interest statement
The authors of declare no conflict of interest.
Figures
References
-
- Saccò M, White NE, Harrod C, Salazar G, Aguilar P, Cubillos CF, Meredith K, Baxter BK, Oren A, Anufriieva E, Shadrin N, Marambio-Alfaro Y, Bravo-Naranjo V, Allentoft ME. 2021. Salt to conserve: a review on the ecology and preservation of hypersaline ecosystems. Biol Rev Camb Philos Soc 96:2828–2850. doi:10.1111/brv.12780 - DOI - PubMed
-
- DasSarma S, Arora P. 2001. Halophiles. e LS.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

