XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1
- PMID: 38377995
- PMCID: PMC10948033
- DOI: 10.1016/j.chom.2024.01.014
XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1
Abstract
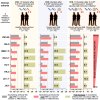
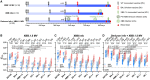
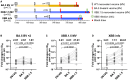
COVID-19 vaccines have recently been updated to specifically encode or contain the spike protein of the SARS-CoV-2 XBB.1.5 subvariant, but their immunogenicity in humans has yet to be fully evaluated and reported, particularly against emergent viruses that are rapidly expanding. We now report that administration of an updated monovalent mRNA vaccine booster (XBB.1.5 MV) to previously uninfected individuals boosted serum virus-neutralizing antibodies significantly against not only XBB.1.5 (27.0-fold increase) and EG.5.1 (27.6-fold increase) but also key emerging viruses such as HV.1, HK.3, JD.1.1, and JN.1 (13.3- to 27.4-fold increase). Individuals previously infected by an Omicron subvariant had the highest overall serum neutralizing titers (ID50 1,504-22,978) against all viral variants tested. While immunological imprinting was still evident with the updated vaccines, it was not nearly as severe as observed with the previously authorized bivalent BA.5 vaccine. Our findings strongly support the official recommendation to widely apply the updated COVID-19 vaccines.
Keywords: COVID-19; HK.3; HV.1; JD.1.1; JN.1; Omicron subvariants; SARS-CoV-2; XBB.1.5 monovalent mRNA vaccine; immunological imprinting; serum neutralization.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.D.H. co-founded TaiMed Biologics and RenBio, serves as a consultant for WuXi Biologics and Brii Biosciences, and is a board director at Vicarious Surgical. A.G. served as a member of the scientific advisory board for Janssen Pharmaceuticals.
Figures


References
-
- WHO . 2023. Statement on the fifteenth meeting of the IHR (2005) Emergency Committee on the COVID-19 pandemic.https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meet...
-
- Wang Q., Guo Y., Liu L., Schwanz L.T., Li Z., Nair M.S., Ho J., Zhang R.M., Iketani S., Yu J., et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature. 2023;624:639–644. - PubMed
-
- Wang Q., Guo Y., Zhang R.M., Ho J., Mohri H., Valdez R., Manthei D.M., Gordon A., Liu L., Ho D.D. Antibody neutralisation of emerging SARS-CoV-2 subvariants: EG.5.1 and XBC.1.6. Lancet Infect. Dis. 2023;23:e397–e398. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

