Synthetically derived BiAux modulates auxin co-receptor activity to stimulate lateral root formation
- PMID: 38378170
- PMCID: PMC11142373
- DOI: 10.1093/plphys/kiae090
Synthetically derived BiAux modulates auxin co-receptor activity to stimulate lateral root formation
Abstract
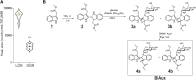
The root system plays an essential role in plant growth and adaptation to the surrounding environment. The root clock periodically specifies lateral root prebranch sites (PBS), where a group of pericycle founder cells (FC) is primed to become lateral root founder cells and eventually give rise to lateral root primordia or lateral roots (LRs). This clock-driven organ formation process is tightly controlled by modulation of auxin content and signaling. Auxin perception entails the physical interaction of TRANSPORT INHIBITOR RESPONSE 1 (TIR1) or AUXIN SIGNALING F-BOX (AFBs) proteins with AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) repressors to form a co-receptor system. Despite the apparent simplicity, the understanding of how specific auxin co-receptors are assembled remains unclear. We identified the compound bis-methyl auxin conjugated with N-glucoside, or BiAux, in Arabidopsis (Arabidopsis thaliana) that specifically induces the formation of PBS and the emergence of LR, with a slight effect on root elongation. Docking analyses indicated that BiAux binds to F-box proteins, and we showed that BiAux function depends on TIR1 and AFB2 F-box proteins and AUXIN RESPONSE FACTOR 7 activity, which is involved in FC specification and LR formation. Finally, using a yeast (Saccharomyces cerevisiae) heterologous expression system, we showed that BiAux favors the assemblage of specific co-receptors subunits involved in LR formation and enhances AUXIN/INDOLE-3-ACETIC ACID 28 protein degradation. These results indicate that BiAux acts as an allosteric modulator of specific auxin co-receptors. Therefore, BiAux exerts a fine-tune regulation of auxin signaling aimed to the specific formation of LR among the many development processes regulated by auxin.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor.Plant J. 2012 May;70(3):492-500. doi: 10.1111/j.1365-313X.2011.04885.x. Epub 2012 Jan 16. Plant J. 2012. PMID: 22171938 Free PMC article.
-
A Molecular Framework for the Control of Adventitious Rooting by TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis.Mol Plant. 2019 Nov 4;12(11):1499-1514. doi: 10.1016/j.molp.2019.09.001. Epub 2019 Sep 11. Mol Plant. 2019. PMID: 31520787
-
Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB.Plant Cell Physiol. 2014 Aug;55(8):1450-9. doi: 10.1093/pcp/pcu077. Epub 2014 May 31. Plant Cell Physiol. 2014. PMID: 24880779
-
Unresolved roles of Aux/IAA proteins in auxin responses.Physiol Plant. 2025 Mar-Apr;177(2):e70221. doi: 10.1111/ppl.70221. Physiol Plant. 2025. PMID: 40265222 Free PMC article. Review.
-
26S Proteasome: Hunter and Prey in Auxin Signaling.Trends Plant Sci. 2016 Jul;21(7):546-548. doi: 10.1016/j.tplants.2016.05.007. Epub 2016 May 27. Trends Plant Sci. 2016. PMID: 27246455 Review.
Cited by
-
Two auxins are better than one: BiAux joins forces with auxin to enhance lateral root formation.Plant Physiol. 2024 May 31;195(2):1114-1116. doi: 10.1093/plphys/kiae132. Plant Physiol. 2024. PMID: 38445801 Free PMC article. No abstract available.
References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

