SNVstory: inferring genetic ancestry from genome sequencing data
- PMID: 38378494
- PMCID: PMC10877842
- DOI: 10.1186/s12859-024-05703-y
SNVstory: inferring genetic ancestry from genome sequencing data
Abstract
Background: Genetic ancestry, inferred from genomic data, is a quantifiable biological parameter. While much of the human genome is identical across populations, it is estimated that as much as 0.4% of the genome can differ due to ancestry. This variation is primarily characterized by single nucleotide variants (SNVs), which are often unique to specific genetic populations. Knowledge of a patient's genetic ancestry can inform clinical decisions, from genetic testing and health screenings to medication dosages, based on ancestral disease predispositions. Nevertheless, the current reliance on self-reported ancestry can introduce subjectivity and exacerbate health disparities. While genomic sequencing data enables objective determination of a patient's genetic ancestry, existing approaches are limited to ancestry inference at the continental level.
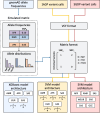
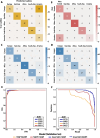
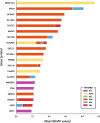
Results: To address this challenge, and create an objective, measurable metric of genetic ancestry we present SNVstory, a method built upon three independent machine learning models for accurately inferring the sub-continental ancestry of individuals. We also introduce a novel method for simulating individual samples from aggregate allele frequencies from known populations. SNVstory includes a feature-importance scheme, unique among open-source ancestral tools, which allows the user to track the ancestral signal broadcast by a given gene or locus. We successfully evaluated SNVstory using a clinical exome sequencing dataset, comparing self-reported ethnicity and race to our inferred genetic ancestry, and demonstrate the capability of the algorithm to estimate ancestry from 36 different populations with high accuracy.
Conclusions: SNVstory represents a significant advance in methods to assign genetic ancestry, opening the door to ancestry-informed care. SNVstory, an open-source model, is packaged as a Docker container for enhanced reliability and interoperability. It can be accessed from https://github.com/nch-igm/snvstory .
Keywords: Genetic ancestry prediction; Genetic variation; Machine learning; Model interpretation; Personalized medicine.
© 2024. The Author(s).
Conflict of interest statement
No competing interests: AEB, AR, DC, JBG, and PW. ERM: Qiagen N.V., supervisory board member, honorarium, and stock-based compensation. Singular Genomics Systems, Inc., board of directors, honorarium, and stock-based compensation.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

