Low-dose hypomethylating agents cooperate with ferroptosis inducers to enhance ferroptosis by regulating the DNA methylation-mediated MAGEA6-AMPK-SLC7A11-GPX4 signaling pathway in acute myeloid leukemia
- PMID: 38378601
- PMCID: PMC10877917
- DOI: 10.1186/s40164-024-00489-4
Low-dose hypomethylating agents cooperate with ferroptosis inducers to enhance ferroptosis by regulating the DNA methylation-mediated MAGEA6-AMPK-SLC7A11-GPX4 signaling pathway in acute myeloid leukemia
Abstract
Background: Ferroptosis is a new form of nonapoptotic and iron-dependent type of cell death. Glutathione peroxidase-4 (GPX4) plays an essential role in anti-ferroptosis by reducing lipid peroxidation. Although acute myeloid leukemia (AML) cells, especially relapsed and refractory (R/R)-AML, present high GPX4 levels and enzyme activities, pharmacological inhibition of GPX4 alone has limited application in AML. Thus, whether inhibition of GPX4 combined with other therapeutic reagents has effective application in AML is largely unknown.
Methods: Lipid reactive oxygen species (ROS), malondialdehyde (MDA), and glutathione (GSH) assays were used to assess ferroptosis in AML cells treated with the hypomethylating agent (HMA) decitabine (DAC), ferroptosis-inducer (FIN) RAS-selective lethal 3 (RSL3), or their combination. Combination index (CI) analysis was used to assess the synergistic activity of DAC + RSL3 against AML cells. Finally, we evaluated the synergistic activity of DAC + RSL3 in murine AML and a human R/R-AML-xenografted NSG model in vivo.
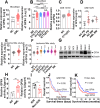
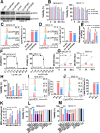
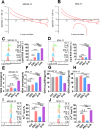
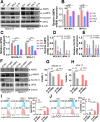
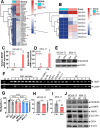
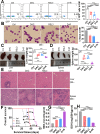
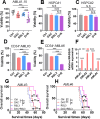
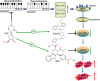
Results: We first assessed GPX4 expression and found that GPX4 levels were higher in AML cells, especially those with MLL rearrangements, than in NCs. Knockdown of GPX4 by shRNA and indirect inhibition of GPX4 enzyme activity by RSL3 robustly induced ferroptosis in AML cells. To reduce the dose of RSL3 and avoid side effects, low doses of DAC (0.5 µM) and RSL3 (0.05 µM) synergistically facilitate ferroptosis by inhibiting the AMP-activated protein kinase (AMPK)-SLC7A11-GPX4 axis. Knockdown of AMPK by shRNA enhanced ferroptosis, and overexpression of SLC7A11 and GPX4 rescued DAC + RSL3-induced anti-leukemogenesis. Mechanistically, DAC increased the expression of MAGEA6 by reducing MAGEA6 promoter hypermethylation. Overexpression of MAGEA6 induced the degradation of AMPK, suggesting that DAC inhibits the AMPK-SLC7A11-GPX4 axis by increasing MAGEA6 expression. In addition, DAC + RSL3 synergistically reduced leukemic burden and extended overall survival compared with either DAC or RSL3 treatment in the MLL-AF9-transformed murine model. Finally, DAC + RSL3 synergistically reduced viability in untreated and R/R-AML cells and extended overall survival in two R/R-AML-xenografted NSG mouse models.
Conclusions: Our study first identify vulnerability to ferroptosis by regulating MAGEA6-AMPK-SLC7A11-GPX4 signaling pathway. Combined treatment with HMAs and FINs provides a potential therapeutic choice for AML patients, especially for R/R-AML.
Keywords: AMPK; Acute myeloid leukemia; Ferroptosis; Glutathione peroxidase-4; Hypomethylating agent.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Integrated transcriptome profiling and in vitro analysis reveals MLN4924's role in inducing ferroptosis in acute myeloid leukemia.Hematology. 2025 Dec;30(1):2497041. doi: 10.1080/16078454.2025.2497041. Epub 2025 May 13. Hematology. 2025. PMID: 40359393
-
[Astragalus polysaccharides induces ferroptosis in ovarian adenocarcinoma cells through Nrf2/SLC7A11/GPX4 signaling pathway].Zhongguo Zhong Yao Za Zhi. 2024 Dec;49(23):6459-6467. doi: 10.19540/j.cnki.cjcmm.20240919.501. Zhongguo Zhong Yao Za Zhi. 2024. PMID: 39805792 Chinese.
-
Targeting NRF2 uncovered an intrinsic susceptibility of acute myeloid leukemia cells to ferroptosis.Exp Hematol Oncol. 2023 May 17;12(1):47. doi: 10.1186/s40164-023-00411-4. Exp Hematol Oncol. 2023. PMID: 37198609 Free PMC article.
-
Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis.Curr Top Microbiol Immunol. 2017;403:143-170. doi: 10.1007/82_2016_508. Curr Top Microbiol Immunol. 2017. PMID: 28204974 Review.
-
Emerging role of glutathione peroxidase 4 in myeloid cell lineage development and acute myeloid leukemia.Cell Mol Biol Lett. 2024 Jul 8;29(1):98. doi: 10.1186/s11658-024-00613-6. Cell Mol Biol Lett. 2024. PMID: 38977956 Free PMC article. Review.
Cited by
-
Unraveling lipid metabolism for acute myeloid leukemia therapy.Curr Opin Hematol. 2025 Mar 1;32(2):77-86. doi: 10.1097/MOH.0000000000000853. Epub 2024 Nov 25. Curr Opin Hematol. 2025. PMID: 39585293 Free PMC article. Review.
-
Targeting oncogenic MAGEA6 sensitizes triple negative breast cancer to doxorubicin through its autophagy and ferroptosis by stabling AMPKα1.Cell Death Discov. 2024 Oct 6;10(1):430. doi: 10.1038/s41420-024-02196-9. Cell Death Discov. 2024. PMID: 39370446 Free PMC article.
-
RSL3 enhances ROS-mediated cell apoptosis of myelodysplastic syndrome cells through MYB/Bcl-2 signaling pathway.Cell Death Dis. 2024 Jul 2;15(7):465. doi: 10.1038/s41419-024-06866-5. Cell Death Dis. 2024. PMID: 38956026 Free PMC article.
-
Ferroptosis: a novel therapeutic warrior in the battle against leukemia.Apoptosis. 2025 Aug;30(7-8):1776-1795. doi: 10.1007/s10495-025-02130-z. Epub 2025 Jun 9. Apoptosis. 2025. PMID: 40488837 Review.
-
Chemokine CXCL13 facilitates anti-FVIII inhibitory antibody development in hemophilia A patients and murine models.Int Immunopharmacol. 2024 Dec 25;143(Pt 2):113472. doi: 10.1016/j.intimp.2024.113472. Epub 2024 Oct 29. Int Immunopharmacol. 2024. PMID: 39471695 Free PMC article.
References
Grants and funding
- 82300194/the National Natural Science Foundation of China
- 81971991/the National Natural Science Foundation of China
- LY19H080003/the Zhejiang Provincial Natural Science Foundation of China
- Y23H080009/the Zhejiang Provincial Natural Science Foundation of China
- z2-2023021/Discipline Cluster of Oncology of Wenzhou Medical University of China
LinkOut - more resources
Full Text Sources

