16S rRNA sequencing analysis of the oral and fecal microbiota in colorectal cancer positives versus colorectal cancer negatives in Iranian population
- PMID: 38378690
- PMCID: PMC10880352
- DOI: 10.1186/s13099-024-00604-0
16S rRNA sequencing analysis of the oral and fecal microbiota in colorectal cancer positives versus colorectal cancer negatives in Iranian population
Erratum in
-
Correction to: 16 S rRNA sequencing analysis of the oral and fecal microbiota in colorectal cancer positives versus colorectal cancer negatives in Iranian population.Gut Pathog. 2024 Mar 19;16(1):15. doi: 10.1186/s13099-024-00607-x. Gut Pathog. 2024. PMID: 38504333 Free PMC article. No abstract available.
Abstract
Background: Colorectal cancer (CRC) poses a significant healthcare challenge, accounting for nearly 6.1% of global cancer cases. Early detection, facilitated by population screening utilizing innovative biomarkers, is pivotal for mitigating CRC incidence. This study aims to scrutinize the fecal and salivary microbiomes of CRC-positive individuals (CPs) in comparison to CRC-negative counterparts (CNs) to enhance early CRC diagnosis through microbial biomarkers.
Material and methods: A total of 80 oral and stool samples were collected from Taleghani Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran, encompassing both CPs and CNs undergoing screening. Microbial profiling was conducted using 16S rRNA sequencing assays, employing the Nextera XT Index Kit on an Illumina NovaSeq platform.
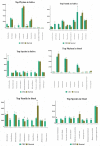
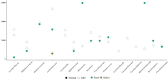
Results: Distinct microbial profiles were observed in saliva and stool samples of CPs, diverging significantly from those of CNs at various taxonomic levels, including phylum, family, and species. Saliva samples from CPs exhibited abundance of Calothrix parietina, Granulicatella adiacens, Rothia dentocariosa, and Rothia mucilaginosa, absent in CNs. Additionally, Lachnospiraceae and Prevotellaceae were markedly higher in CPs' feces, while the Fusobacteria phylum was significantly elevated in CPs' saliva. Conversely, the non-pathogenic bacterium Akkermansia muciniphila exhibited a significant decrease in CPs' fecal samples compared to CNs.
Conclusion: Through meticulous selection of saliva and stool microbes based on Mean Decrease GINI values and employing logistic regression for saliva and support vector machine models for stool, we successfully developed a microbiota test with heightened sensitivity and specificity for early CRC detection.
Keywords: 16S rRNA sequencing; Colorectal cancer; Early detection; Fecal microbiota; Oral microbiota.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests.
Figures


References
Grants and funding
- RIGLD1065/Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- RIGLD1065/Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- Regione Autonoma della Sardegna, legge regionale 12 dicembre 2022, n. 22/UNISS FAR fondi ricercar 2021, 2022 and Fondazione di Sardegna 2017
- Regione Autonoma della Sardegna, legge regionale 12 dicembre 2022, n. 22/UNISS FAR fondi ricercar 2021, 2022 and Fondazione di Sardegna 2017
LinkOut - more resources
Full Text Sources

