Exploiting lung adaptation and phage steering to clear pan-resistant Pseudomonas aeruginosa infections in vivo
- PMID: 38378698
- PMCID: PMC10879199
- DOI: 10.1038/s41467-024-45785-z
Exploiting lung adaptation and phage steering to clear pan-resistant Pseudomonas aeruginosa infections in vivo
Abstract
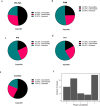
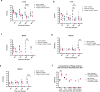
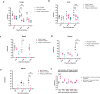
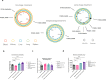
Pseudomonas aeruginosa is a major nosocomial pathogen that causes severe disease including sepsis. Carbapenem-resistant P. aeruginosa is recognised by the World Health Organisation as a priority 1 pathogen, with urgent need for new therapeutics. As such, there is renewed interest in using bacteriophages as a therapeutic. However, the dynamics of treating pan-resistant P. aeruginosa with phage in vivo are poorly understood. Using a pan-resistant P. aeruginosa in vivo infection model, phage therapy displays strong therapeutic potential, clearing infection from the blood, kidneys, and spleen. Remaining bacteria in the lungs and liver displays phage resistance due to limiting phage adsorption. Yet, resistance to phage results in re-sensitisation to a wide range of antibiotics. In this work, we use phage steering in vivo, pre-exposing a pan resistant P. aeruginosa infection with a phage cocktail to re-sensitise bacteria to antibiotics, clearing the infection from all organs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- European Centre for Disease Prevention and Control. Healthcare-associated infections acquired in intensive care units. In: ECDC. Annual epidemiological report for 2017. (ECDC, 2019).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

