A mutational atlas for Parkin proteostasis
- PMID: 38378758
- PMCID: PMC10879094
- DOI: 10.1038/s41467-024-45829-4
A mutational atlas for Parkin proteostasis
Abstract
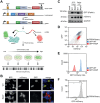
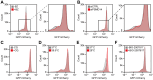
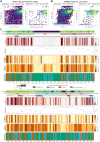
Proteostasis can be disturbed by mutations affecting folding and stability of the encoded protein. An example is the ubiquitin ligase Parkin, where gene variants result in autosomal recessive Parkinsonism. To uncover the pathological mechanism and provide comprehensive genotype-phenotype information, variant abundance by massively parallel sequencing (VAMP-seq) is leveraged to quantify the abundance of Parkin variants in cultured human cells. The resulting mutational map, covering 9219 out of the 9300 possible single-site amino acid substitutions and nonsense Parkin variants, shows that most low abundance variants are proteasome targets and are located within the structured domains of the protein. Half of the known disease-linked variants are found at low abundance. Systematic mapping of degradation signals (degrons) reveals an exposed degron region proximal to the so-called "activation element". This work provides examples of how missense variants may cause degradation either via destabilization of the native protein, or by introducing local signals for degradation.
© 2024. The Author(s).
Conflict of interest statement
K.L.-L. holds stock options in and is a consultant for Peptone Ltd. The remaining authors declare no competing interest.
Figures



References
-
- Poewe W, et al. Parkinson disease. Nat. Rev. Dis. Prim. 2017;3:1–21. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

