Clinical and biomarker results from a phase II trial of combined cabozantinib and durvalumab in patients with chemotherapy-refractory colorectal cancer (CRC): CAMILLA CRC cohort
- PMID: 38378868
- PMCID: PMC10879200
- DOI: 10.1038/s41467-024-45960-2
Clinical and biomarker results from a phase II trial of combined cabozantinib and durvalumab in patients with chemotherapy-refractory colorectal cancer (CRC): CAMILLA CRC cohort
Abstract
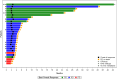
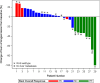
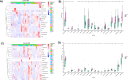
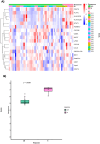
CAMILLA is a basket trial (NCT03539822) evaluating cabozantinib plus the ICI durvalumab in chemorefractory gastrointestinal cancer. Herein, are the phase II colorectal cohort results. 29 patients were evaluable. 100% had confirmed pMMR/MSS tumors. Primary endpoint was met with ORR of 27.6% (95% CI 12.7-47.2%). Secondary endpoints of 4-month PFS rate was 44.83% (95% CI 26.5-64.3%); and median OS was 9.1 months (95% CI 5.8-20.2). Grade≥3 TRAE occurred in 39%. In post-hoc analysis of patients with RAS wild type tumors, ORR was 50% and median PFS and OS were 6.3 and 21.5 months respectively. Exploratory spatial transcriptomic profiling of pretreatment tumors showed upregulation of VEGF and MET signaling, increased extracellular matrix activity and preexisting anti-tumor immune responses coexisting with immune suppressive features like T cell migration barriers in responders versus non-responders. Cabozantinib plus durvalumab demonstrated anti-tumor activity, manageable toxicity, and have led to the activation of the phase III STELLAR-303 trial.
© 2024. The Author(s).
Conflict of interest statement
A.S. reports a leadership role with Autem therapeutics, Exelixis, KAHR medical and Bristol-Myers Squibb; consulting or advisory board role with AstraZeneca, Bristol-Myers Squibb, Merck, Exelixis, Pfizer, Xilio therapeutics, Taiho, Amgen, Autem therapeutics, KAHR medical, and Daiichi Sankyo; institutional research funding from AstraZeneca, Bristol-Myers Squibb, Merck, Clovis, Exelixis, Actuate therapeutics, Incyte Corporation, Daiichi Sankyo, Five prime therapeutics, Amgen, Innovent biologics, Dragonfly therapeutics, Oxford Biotherapeutics, Arcus therapeutics, and KAHR medical; and participation as a data safety monitoring board chair for Arcus therapeutics. R.A. reports research grants (to institution) from AstraZeneca, Bayor, Merck, Bristol Myers Squibb, Exelixis, and Eureka Therapeutics and stock ownership in Actinium Pharmaceuticals and Seagen. A.K. reports research funding (to institution) from Astellas, Tesaro, and Bavarian Nordic. J.B. reports research grants (to institution) from Astellas, Chungchun Intellicrown, Impact Therapeutics, Poseida Therapeutics, Takeda Oncology, and Genome Co. and consultant fees (to institution) from Sanofi. S.W. reports stock/other ownership in Horizon Therapeutics, Iovance Biotherapeutics, and Merus and institutional research funding from Aleon Pharma, Astellas Pharma, Bayer Health, Bristol Myers Squibb, Daiichi Sankyo, EMD Serono, Merck Serono, Nektar, Novartis, Pharmacyclics, AbbVie, Regeneron, Rogosin Institute, Sanofi, Seagen, and Sotio. A.K.G. reports research funding from Predicine and VITRAC Therapeutics, is a co-founder of Sinochips Diagnostics, and serves as a scientific advisory board member to Biovica, Clara Biotech, and Sinochips Diagnostics. The remaining authors report no conflicts of interest.
Figures



References
-
- Diaz LA, Jr, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022;23:659–670. doi: 10.1016/S1470-2045(22)00197-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

