Genetic characteristics of SARS-CoV-2 virus variants observed upon three waves of the COVID-19 pandemic in Ukraine between February 2021-January 2022
- PMID: 38380034
- PMCID: PMC10877268
- DOI: 10.1016/j.heliyon.2024.e25618
Genetic characteristics of SARS-CoV-2 virus variants observed upon three waves of the COVID-19 pandemic in Ukraine between February 2021-January 2022
Abstract
The aim: of our study was to identify and characterize the SARS-CoV-2 variants in COVID-19 patients' samples collected from different regions of Ukraine to determine the relationship between SARS-CoV-2 phylogenetics and COVID-19 epidemiology.
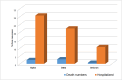
Patients and methods: Samples were collected from COVID-19 patients during 2021 and the beginning of 2022 (401 patients). The SARS-CoV-2 genotyping was performed by parallel whole genome sequencing.
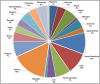
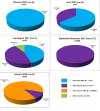
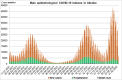
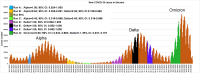
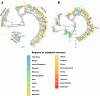
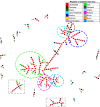
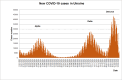
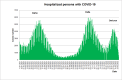
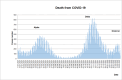
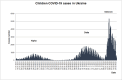
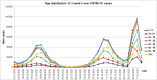
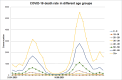
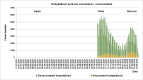
Results: The obtained SARS-CoV-2 genotypes showed that three waves of the COVID-19 pandemic in Ukraine were represented by three main variants of concern (VOC), named Alpha, Delta and Omicron; each VOC successfully replaced the earlier variant. The VOC Alpha strain was presented by one B.1.1.7 lineage, while VOC Delta showed a spectrum of 25 lineages that had different prevalence in 19 investigated regions of Ukraine. The VOC Omicron in the first half of the pandemic was represented by 13 lines that belonged to two different clades representing B.1 and B.2 Omicron strains. Each of the three epidemic waves (VOC Alpha, Delta, and Omicron) demonstrated their own course of disease, associated with genetic changes in the SARS-CoV-2 genome. The observed epidemiological features are associated with the genetic characteristics of the different VOCs, such as point mutations, deletions and insertions in the viral genome. A phylogenetic and transmission analysis showed the different mutation rates; there were multiple virus sources with a limited distribution between regions.
Conclusions: The evolution of SARS-CoV-2 virus and high levels of morbidity due to COVID-19 are still registered in the world. Observed multiple virus sourses with the limited distribution between regions indicates the high efficiency of the anti-epidemic policy pursued by the Ministry of Health of Ukraine to prevent the spread of the epidemic, despite the low level of vaccination of the Ukrainian population.
Keywords: COVID-19; Delta; NGS; Omicron; SARS-CoV-2 genotyping; VOC Alpha.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Molecular epidemiology of SARS-CoV-2 in Northern South Africa: wastewater surveillance from January 2021 to May 2022.Front Public Health. 2023 Dec 19;11:1309869. doi: 10.3389/fpubh.2023.1309869. eCollection 2023. Front Public Health. 2023. PMID: 38174083 Free PMC article.
-
Emergency SARS-CoV-2 Variants of Concern: Novel Multiplex Real-Time RT-PCR Assay for Rapid Detection and Surveillance.Microbiol Spectr. 2022 Feb 23;10(1):e0251321. doi: 10.1128/spectrum.02513-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196812 Free PMC article.
-
Update on SARS-CoV-2 Omicron Variant of Concern and Its Peculiar Mutational Profile.Microbiol Spectr. 2022 Apr 27;10(2):e0273221. doi: 10.1128/spectrum.02732-21. Epub 2022 Mar 30. Microbiol Spectr. 2022. PMID: 35352942 Free PMC article.
-
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
Cited by
-
Genomic analysis of three SARS-CoV-2 waves in west Sumatra: Evolutionary dynamics and variant clustering.Heliyon. 2024 Jul 9;10(14):e34365. doi: 10.1016/j.heliyon.2024.e34365. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39108880 Free PMC article.
-
Genomic Epidemiology of SARS-CoV-2 in Ukraine from May 2022 to March 2024 Reveals Omicron Variant Dynamics.Viruses. 2025 Jul 17;17(7):1000. doi: 10.3390/v17071000. Viruses. 2025. PMID: 40733617 Free PMC article.
-
Genomic epidemiology and evolutionary dynamics of the Omicron variant of SARS-CoV-2 during the fifth wave of COVID-19 in Pakistan.Front Cell Infect Microbiol. 2024 Oct 22;14:1484637. doi: 10.3389/fcimb.2024.1484637. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39502171 Free PMC article.
-
Evaluation of Genomic Surveillance of SARS-CoV-2 Virus Isolates and Comparison of Mutational Spectrum of Variants in Bangladesh.Viruses. 2025 Jan 27;17(2):182. doi: 10.3390/v17020182. Viruses. 2025. PMID: 40006937 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

