Gene inversion led to the emergence of brackish archaeal heterotrophs in the aftermath of the Cryogenian Snowball Earth
- PMID: 38380056
- PMCID: PMC10877094
- DOI: 10.1093/pnasnexus/pgae057
Gene inversion led to the emergence of brackish archaeal heterotrophs in the aftermath of the Cryogenian Snowball Earth
Abstract
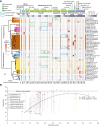
Land-ocean interactions greatly impact the evolution of coastal life on earth. However, the ancient geological forces and genetic mechanisms that shaped evolutionary adaptations and allowed microorganisms to inhabit coastal brackish waters remain largely unexplored. In this study, we infer the evolutionary trajectory of the ubiquitous heterotrophic archaea Poseidoniales (Marine Group II archaea) presently occurring across global aquatic habitats. Our results show that their brackish subgroups had a single origination, dated to over 600 million years ago, through the inversion of the magnesium transport gene corA that conferred osmotic-stress tolerance. The subsequent loss and gain of corA were followed by genome-wide adjustment, characterized by a general two-step mode of selection in microbial speciation. The coastal family of Poseidoniales showed a rapid increase in the evolutionary rate during and in the aftermath of the Cryogenian Snowball Earth (∼700 million years ago), possibly in response to the enhanced phosphorus supply and the rise of algae. Our study highlights the close interplay between genetic changes and ecosystem evolution that boosted microbial diversification in the Neoproterozoic continental margins, where the Cambrian explosion of animals soon followed.
Keywords: Snowball Earth; archaeal evolution; brackish microorganisms; gene inversion; osmotic stress.
© The Author(s) 2024. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures



Similar articles
-
Diversity, distribution, and functional potentials of magroviruses from marine and brackish waters.Front Microbiol. 2023 Apr 21;14:1151034. doi: 10.3389/fmicb.2023.1151034. eCollection 2023. Front Microbiol. 2023. PMID: 37152742 Free PMC article.
-
Cryogenian Glacial Habitats as a Plant Terrestrialisation Cradle - The Origin of the Anydrophytes and Zygnematophyceae Split.Front Plant Sci. 2022 Jan 27;12:735020. doi: 10.3389/fpls.2021.735020. eCollection 2021. Front Plant Sci. 2022. PMID: 35154170 Free PMC article.
-
Cryogenian evolution of stigmasteroid biosynthesis.Sci Adv. 2017 Sep 20;3(9):e1700887. doi: 10.1126/sciadv.1700887. eCollection 2017 Sep. Sci Adv. 2017. PMID: 28948220 Free PMC article. Review.
-
Cryoconite pans on Snowball Earth: supraglacial oases for Cryogenian eukaryotes?Geobiology. 2016 Nov;14(6):531-542. doi: 10.1111/gbi.12191. Epub 2016 Jul 15. Geobiology. 2016. PMID: 27422766
-
Salinity tolerances and use of saline environments by freshwater turtles: implications of sea level rise.Biol Rev Camb Philos Soc. 2018 Aug;93(3):1634-1648. doi: 10.1111/brv.12410. Epub 2018 Mar 25. Biol Rev Camb Philos Soc. 2018. PMID: 29575680 Review.
Cited by
-
Multiple Environmental Factors Shaping Hopanoid-Producing Microbes Across Different Ecosystems.Microorganisms. 2025 May 28;13(6):1250. doi: 10.3390/microorganisms13061250. Microorganisms. 2025. PMID: 40572137 Free PMC article.
References
Associated data
LinkOut - more resources
Full Text Sources

