The diversification and potential function of microbiome in sediment-water interface of methane seeps in South China Sea
- PMID: 38380093
- PMCID: PMC10878133
- DOI: 10.3389/fmicb.2024.1287147
The diversification and potential function of microbiome in sediment-water interface of methane seeps in South China Sea
Abstract
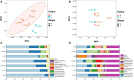
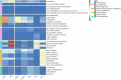
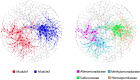
The sediment-water interfaces of cold seeps play important roles in nutrient transportation between seafloor and deep-water column. Microorganisms are the key actors of biogeochemical processes in this interface. However, the knowledge of the microbiome in this interface are limited. Here we studied the microbial diversity and potential metabolic functions by 16S rRNA gene amplicon sequencing at sediment-water interface of two active cold seeps in the northern slope of South China Sea, Lingshui and Site F cold seeps. The microbial diversity and potential functions in the two cold seeps are obviously different. The microbial diversity of Lingshui interface areas, is found to be relatively low. Microbes associated with methane consumption are enriched, possibly due to the large and continuous eruptions of methane fluids. Methane consumption is mainly mediated by aerobic oxidation and denitrifying anaerobic methane oxidation (DAMO). The microbial diversity in Site F is higher than Lingshui. Fluids from seepage of Site F are mitigated by methanotrophic bacteria at the cyclical oxic-hypoxic fluctuating interface where intense redox cycling of carbon, sulfur, and nitrogen compounds occurs. The primary modes of microbial methane consumption are aerobic methane oxidation, along with DAMO, sulfate-dependent anaerobic methane oxidation (SAMO). To sum up, anaerobic oxidation of methane (AOM) may be underestimated in cold seep interface microenvironments. Our findings highlight the significance of AOM and interdependence between microorganisms and their environments in the interface microenvironments, providing insights into the biogeochemical processes that govern these unique ecological systems.
Keywords: DAMO; cold seeps; methane metabolism; oxic-hypoxic shifting; sediment-water interface.
Copyright © 2024 Fu, Liu, Wang, Lian, Cao, Wang, Sun, Wang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Cold Seeps on the Passive Northern U.S. Atlantic Margin Host Globally Representative Members of the Seep Microbiome with Locally Dominant Strains of Archaea.Appl Environ Microbiol. 2022 Jun 14;88(11):e0046822. doi: 10.1128/aem.00468-22. Epub 2022 May 24. Appl Environ Microbiol. 2022. PMID: 35607968 Free PMC article.
-
Biogeographic distributions of microbial communities associated with anaerobic methane oxidation in the surface sediments of deep-sea cold seeps in the South China Sea.Front Microbiol. 2022 Dec 23;13:1060206. doi: 10.3389/fmicb.2022.1060206. eCollection 2022. Front Microbiol. 2022. PMID: 36620029 Free PMC article.
-
Anaerobic methane oxidation coupled to denitrification is an important potential methane sink in deep-sea cold seeps.Sci Total Environ. 2020 Dec 15;748:142459. doi: 10.1016/j.scitotenv.2020.142459. Epub 2020 Sep 23. Sci Total Environ. 2020. PMID: 33113688
-
Unexpected genetic and microbial diversity for arsenic cycling in deep sea cold seep sediments.NPJ Biofilms Microbiomes. 2023 Mar 29;9(1):13. doi: 10.1038/s41522-023-00382-8. NPJ Biofilms Microbiomes. 2023. PMID: 36991068 Free PMC article. Review.
-
Genomic Insights into Niche Partitioning across Sediment Depth among Anaerobic Methane-Oxidizing Archaea in Global Methane Seeps.mSystems. 2023 Apr 27;8(2):e0117922. doi: 10.1128/msystems.01179-22. Epub 2023 Mar 16. mSystems. 2023. PMID: 36927099 Free PMC article. Review.
Cited by
-
Biotic Interaction Underpins the Assembly Processes of the Bacterial Community Across the Sediment-Water Interface in a Subalpine Lake.Microorganisms. 2024 Nov 25;12(12):2418. doi: 10.3390/microorganisms12122418. Microorganisms. 2024. PMID: 39770621 Free PMC article.
References
-
- Ai C., Liang G., Wang X., Sun J., He P., Zhou W. (2017). A distinctive root-inhabiting denitrifying community with high N2O/(N2O+N2) product ratio. Soil Biol. Biochem. 109 118–123.
-
- Bastian M., Heymann S., Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3, 361–362.
LinkOut - more resources
Full Text Sources

