Effect of TraN key residues involved in DNA binding on pIP501 transfer rates in Enterococcus faecalis
- PMID: 38380428
- PMCID: PMC10877727
- DOI: 10.3389/fmolb.2024.1268647
Effect of TraN key residues involved in DNA binding on pIP501 transfer rates in Enterococcus faecalis
Abstract
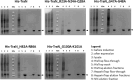
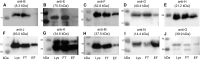
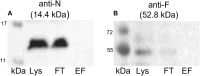
Conjugation is a major mechanism that facilitates the exchange of antibiotic resistance genes among bacteria. The broad-host-range Inc18 plasmid pIP501 harbors 15 genes that encode for a type IV secretion system (T4SS). It is a membrane-spanning multiprotein complex formed between conjugating donor and recipient cells. The penultimate gene of the pIP501 operon encodes for the cytosolic monomeric protein TraN. This acts as a transcriptional regulator by binding upstream of the operon promotor, partially overlapping with the origin of transfer. Additionally, TraN regulates traN and traO expression by binding upstream of the PtraNO promoter. This study investigates the impact of nine TraN amino acids involved in binding to pIP501 DNA through site-directed mutagenesis by exchanging one to three residues by alanine. For three traN variants, complementation of the pIP501∆traN knockout resulted in an increase of the transfer rate by more than 1.5 orders of magnitude compared to complementation of the mutant with native traN. Microscale thermophoresis (MST) was used to assess the binding affinities of three TraN double-substituted variants and one triple-substituted variant to its cognate pIP501 double-stranded DNA. The MST data strongly correlated with the transfer rates obtained by biparental mating assays in Enterococcus faecalis. The TraN variants TraN_R23A-N24A-Q28A, TraN_H82A-R86A, and TraN_G100A-K101A not only exhibited significantly lower DNA binding affinities but also, upon complementation of the pIP501∆traN knockout, resulted in the highest pIP501 transfer rates. This confirms the important role of the TraN residues R23, N24, Q28, H82, R86, G100, and K101 in downregulating pIP501 transfer. Although TraN is not part of the mating pair formation complex, TraE, TraF, TraH, TraJ, TraK, and TraM were coeluted with TraN in a pull-down. Moreover, TraN homologs are present not only in Inc18 plasmids but also in RepA_N and Rep_3 family plasmids, which are frequently found in enterococci, streptococci, and staphylococci. This points to a widespread role of this repressor in conjugative plasmid transfer among Firmicutes.
Keywords: DNA binding proteins; Enterococcus faecalis; antibiotic resistance; conjugative transfer; pIP501; transfer regulator; type IV secretion system.
Copyright © 2024 Michaelis, Berger, Kuhlmann, Ghulam, Petrowitsch, Besora Vecino, Gesslbauer, Pavkov-Keller, Keller and Grohmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were editorial board members of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Figures





Similar articles
-
DNA-Binding Proteins Regulating pIP501 Transfer and Replication.Front Mol Biosci. 2016 Aug 11;3:42. doi: 10.3389/fmolb.2016.00042. eCollection 2016. Front Mol Biosci. 2016. PMID: 27563645 Free PMC article. Review.
-
Structure of the double-stranded DNA-binding type IV secretion protein TraN from Enterococcus.Acta Crystallogr D Biol Crystallogr. 2014 Sep;70(Pt 9):2376-89. doi: 10.1107/S1399004714014187. Epub 2014 Aug 29. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 25195751
-
Small Things Matter: The 11.6-kDa TraB Protein is Crucial for Antibiotic Resistance Transfer Among Enterococci.Front Mol Biosci. 2022 Apr 25;9:867136. doi: 10.3389/fmolb.2022.867136. eCollection 2022. Front Mol Biosci. 2022. PMID: 35547396 Free PMC article.
-
VirB8-like protein TraH is crucial for DNA transfer in Enterococcus faecalis.Sci Rep. 2016 Apr 22;6:24643. doi: 10.1038/srep24643. Sci Rep. 2016. PMID: 27103580 Free PMC article.
-
Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms.Plasmid. 2018 Sep;99:11-21. doi: 10.1016/j.plasmid.2018.06.001. Epub 2018 Jun 19. Plasmid. 2018. PMID: 29932966 Review.
Cited by
-
Interactive 3D Objects Enhance Scientific Communication of Structural Data.Chembiochem. 2025 Mar 15;26(6):e202500036. doi: 10.1002/cbic.202500036. Epub 2025 Feb 20. Chembiochem. 2025. PMID: 39976303 Free PMC article. Review.
-
Comparative genomic analysis of ten Elizabethkingia anophelis isolated from clinical patients in China.Microbiol Spectr. 2025 Jan 7;13(1):e0178024. doi: 10.1128/spectrum.01780-24. Epub 2024 Nov 29. Microbiol Spectr. 2025. PMID: 39612476 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

