RECQL4 Inhibits Radiation-Induced Tumor Immune Awakening via Suppressing the cGAS-STING Pathway in Hepatocellular Carcinoma
- PMID: 38381090
- PMCID: PMC11040365
- DOI: 10.1002/advs.202308009
RECQL4 Inhibits Radiation-Induced Tumor Immune Awakening via Suppressing the cGAS-STING Pathway in Hepatocellular Carcinoma
Abstract
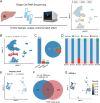
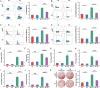
Many patients with hepatocellular carcinoma (HCC) respond poorly to radiotherapy despite remarkable advances in treatment. A deeper insight into the mechanism of sensitivity of HCC to this therapy is urgently required. It is demonstrated that RECQL4 is upregulated in the malignant cells of patients with HCC. Elevated RECQL4 levels reduce the sensitivity of HCC to radiotherapy by repairing radiation-induced double-stranded DNA (dsDNA) fragments. Mechanistically, the inhibitory effect of RECQL4 on radiotherapy is due to the reduced recruitment of dendritic cells and CD8+ T cells in the tumor microenvironment (TME). RECQL4 disrupts the radiation-induced transformation of the TME into a tumoricidal niche by inhibiting the cGAS-STING pathway in dendritic cells. Knocking out STING in dendritic cells can block the impact of RECQL4 on HCC radiosensitivity. Notably, high RECQL4 expressions in HCC is significantly associated with poor prognosis in multiple independent cohorts. In conclusion, this study highlights how HCC-derived RECQL4 disrupts cGAS-STING pathway activation in dendritic cells through DNA repair, thus reducing the radiosensitivity of HCC. These findings provide new perspectives on the clinical treatment of HCC.
Keywords: DNA repair; RecQ‐Like Helicase 4; cGAS‐STING pathway; hepatocellular carcinoma; tumor microenvironment.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J Clin 2021, 71, 209. - PubMed
-
- Riaz A., JAMA, J. Am. Med. Assoc. 2010, 303, 1062.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
