Identification of a novel and plant height-independent QTL for coleoptile length in barley and validation of its effect using near isogenic lines
- PMID: 38381194
- PMCID: PMC10881613
- DOI: 10.1007/s00122-024-04561-9
Identification of a novel and plant height-independent QTL for coleoptile length in barley and validation of its effect using near isogenic lines
Abstract
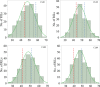
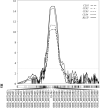
This study reported the identification and validation of novel QTL conferring coleoptile length in barley and predicted candidate genes underlying the largest effect QTL based on orthologous analysis and comparison of the whole genome assemblies for both parental genotypes of the mapping population. Coleoptile length (CL) is one of the most important agronomic traits in cereal crops due to its direct influence on the optimal depth for seed sowing which facilitates better seedling establishment. Varieties with longer coleoptiles are preferred in drought-prone areas where less moisture maintains at the top layer of the soil. Compared to wheat, genetic study on coleoptile length is limited in barley. Here, we reported a study on detecting the genomic regions associated with CL in barley by assessing a population consisting of 201 recombinant inbred lines. Four putative QTL conferring CL were consistently identified on chromosomes 1H, 5H, 6H, and 7H in each of the trials conducted. Of these QTL, the two located on chromosomes 5H and 6H (designated as Qcl.caf-5H and Qcl.caf-6H) are likely novel and Qcl.caf-5H showed the most significant effect explaining up to 30.9% of phenotypic variance with a LOD value of 15.1. To further validate the effect of this putative QTL, five pairs of near isogenic lines (NILs) were then developed and assessed. Analysis of the NILs showed an average difference of 21.0% in CL between the two isolines. Notably, none of the other assessed morphological characteristics showed consistent differences between the two isolines for each pair of the NILs. Candidate genes underlying the Qcl.caf-5H locus were also predicted by employing orthologous analysis and comparing the genome assemblies for both parental genotypes of the mapping population in the present study. Taken together, these findings expand our understanding on genetic basis of CL and will be indicative for further gene cloning and functional analysis underly this locus in barley.
© 2024. Crown.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Unveiling the genetic architecture of barley embryo: QTL mapping, candidate genes identification and its relationship with kernel size and early vigour.Theor Appl Genet. 2025 Jan 23;138(1):32. doi: 10.1007/s00122-025-04817-y. Theor Appl Genet. 2025. PMID: 39843841 Free PMC article.
-
Genome-wide association mapping reveals novel genes associated with coleoptile length in a worldwide collection of barley.BMC Plant Biol. 2020 Jul 22;20(1):346. doi: 10.1186/s12870-020-02547-5. BMC Plant Biol. 2020. PMID: 32698771 Free PMC article.
-
Fine mapping of a Fusarium crown rot resistant locus on chromosome arm 6HL in barley by exploiting near isogenic lines, transcriptome profiling, and a large near isogenic line-derived population.Theor Appl Genet. 2023 May 26;136(6):137. doi: 10.1007/s00122-023-04387-x. Theor Appl Genet. 2023. PMID: 37233855 Free PMC article.
-
Leaf thickness of barley: genetic dissection, candidate genes prediction and its relationship with yield-related traits.Theor Appl Genet. 2022 Jun;135(6):1843-1854. doi: 10.1007/s00122-022-04076-1. Epub 2022 Mar 29. Theor Appl Genet. 2022. PMID: 35348823
-
Simultaneous selection of major and minor genes: use of QTL to increase selection efficiency of coleoptile length of wheat (Triticum aestivum L.).Theor Appl Genet. 2009 Jun;119(1):65-74. doi: 10.1007/s00122-009-1017-2. Epub 2009 Apr 10. Theor Appl Genet. 2009. PMID: 19360392
Cited by
-
High-throughput phenotyping of wheat root angle and coleoptile length at different temperatures using 3D-printed equipment.BMC Plant Biol. 2025 Jan 25;25(1):112. doi: 10.1186/s12870-025-06120-w. BMC Plant Biol. 2025. PMID: 39863883 Free PMC article.
-
Unveiling the genetic architecture of barley embryo: QTL mapping, candidate genes identification and its relationship with kernel size and early vigour.Theor Appl Genet. 2025 Jan 23;138(1):32. doi: 10.1007/s00122-025-04817-y. Theor Appl Genet. 2025. PMID: 39843841 Free PMC article.
References
-
- Ahmadi-Ochtapeh H, Soltanloo H, Ramezanpour SS, et al. QTL mapping for salt tolerance in barley at seedling growth stage. Biol Plant. 2015;59:283–290. doi: 10.1007/s10535-015-0496-z. - DOI
-
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. doi: 10.18637/jss.v067.i01. - DOI
-
- Brand J, Yaduraju N, Shivakumar B, Murray L. Weed management. In: Yadav SS, McNeil DL, Stevenson PC, editors. Lentil—an ancient crop for modern times. Dordrecht: Springer; 2007. pp. 159–172.
-
- Brown PR, Singleton GR, Tann CR, Mock I. Increasing sowing depth to reduce mouse damage to winter crops. Crop Prot. 2003;22:653–660. doi: 10.1016/S0261-2194(03)00006-1. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

