PABPC1L Induces IDO1 to Promote Tryptophan Metabolism and Immune Suppression in Renal Cell Carcinoma
- PMID: 38382068
- PMCID: PMC11094425
- DOI: 10.1158/0008-5472.CAN-23-2521
PABPC1L Induces IDO1 to Promote Tryptophan Metabolism and Immune Suppression in Renal Cell Carcinoma
Abstract
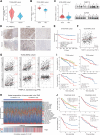
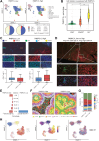
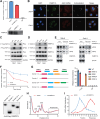
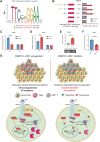
The tumor microenvironment (TME) in renal cell carcinomas (RCC) is marked by substantial immunosuppression and immune resistance despite having extensive T-cell infiltration. Elucidation of the mechanisms underlying immune evasion could help identify therapeutic strategies to boost the efficacy of immune checkpoint blockade (ICB) in RCC. This study uncovered a mechanism wherein the polyadenylate-binding protein PABPC1L modulates indoleamine 2,3-dioxygenase 1 (IDO1), a prospective target for immunotherapy. PABPC1L was markedly upregulated in RCC, and high PABPC1L expression correlated with unfavorable prognosis and resistance to ICB. PABPC1L bolstered tryptophan metabolism by upregulating IDO1, inducing T-cell dysfunction and Treg infiltration. PABPC1L enhanced the stability of JAK2 mRNA, leading to increased JAK2-STAT1 signaling that induced IDO1 expression. Additionally, PABPC1L-induced activation of the JAK2-STAT1 axis created a positive feedback loop to promote PABPC1L transcription. Conversely, loss of PABPC1L diminished IDO1 expression, mitigated cytotoxic T-cell suppression, and enhanced responsiveness to anti-PD-1 therapy in patient-derived xenograft models. These findings reveal the crucial role of PABPC1L in facilitating immune evasion in RCC and indicate that inhibiting PABPC1L could be a potential immunotherapeutic approach in combination with ICB to improve patient outcomes.
Significance: PABPC1L functions as a key factor in renal cell carcinoma immune evasion, enhancing IDO1 and impeding T-cell function, and represents a potential target to enhance the efficacy of immune checkpoint blockade therapy.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures


Similar articles
-
TUBA1C orchestrates the immunosuppressive tumor microenvironment and resistance to immune checkpoint blockade in clear cell renal cell carcinoma.Front Immunol. 2024 Sep 5;15:1457691. doi: 10.3389/fimmu.2024.1457691. eCollection 2024. Front Immunol. 2024. PMID: 39301023 Free PMC article.
-
Tryptophan 2,3-dioxygenase in tumor cells is associated with resistance to immunotherapy in renal cell carcinoma.Cancer Sci. 2021 Mar;112(3):1038-1047. doi: 10.1111/cas.14797. Epub 2021 Jan 25. Cancer Sci. 2021. PMID: 33410234 Free PMC article.
-
EHBP1L1 Drives Immune Evasion in Renal Cell Carcinoma through Binding and Stabilizing JAK1.Adv Sci (Weinh). 2023 Apr;10(11):e2206792. doi: 10.1002/advs.202206792. Epub 2023 Feb 12. Adv Sci (Weinh). 2023. PMID: 36775874 Free PMC article.
-
Indoleamine 2,3-dioxygenase 1 in circumventing checkpoint inhibitor responses: Updated.Int Immunopharmacol. 2023 May;118:110032. doi: 10.1016/j.intimp.2023.110032. Epub 2023 Mar 16. Int Immunopharmacol. 2023. PMID: 36933494 Review.
-
Indoleamine 2,3-Dioxygenase 1: A Promising Therapeutic Target in Malignant Tumor.Front Immunol. 2021 Dec 23;12:800630. doi: 10.3389/fimmu.2021.800630. eCollection 2021. Front Immunol. 2021. PMID: 35003126 Free PMC article. Review.
Cited by
-
Metabolic checkpoints in immune cell reprogramming: rewiring immunometabolism for cancer therapy.Mol Cancer. 2025 Aug 2;24(1):210. doi: 10.1186/s12943-025-02407-6. Mol Cancer. 2025. PMID: 40753443 Free PMC article. Review.
-
Cancer cells accelerate exhaustion of persistently activated mouse CD4+ T cells.Oncoimmunology. 2025 Dec;14(1):2521392. doi: 10.1080/2162402X.2025.2521392. Epub 2025 Jun 19. Oncoimmunology. 2025. PMID: 40536473 Free PMC article.
-
New insights into T cell metabolism in liver cancer: from mechanism to therapy.Cell Death Discov. 2025 Mar 23;11(1):118. doi: 10.1038/s41420-025-02397-w. Cell Death Discov. 2025. PMID: 40122853 Free PMC article. Review.
-
Histone deacetylases 10 as a prognostic biomarker correlates with tumor microenvironment and therapy response in colorectal cancer.World J Gastroenterol. 2025 Jul 14;31(26):108662. doi: 10.3748/wjg.v31.i26.108662. World J Gastroenterol. 2025. PMID: 40678705 Free PMC article.
-
Bioinformatics analysis and experimental verification of the cancer-promoting effect of DHODH in clear cell renal cell carcinoma.Sci Rep. 2024 May 25;14(1):11985. doi: 10.1038/s41598-024-62738-0. Sci Rep. 2024. PMID: 38796629 Free PMC article.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. - PubMed
-
- Williamson SR, Taneja K, Cheng L. Renal cell carcinoma staging: pitfalls, challenges, and updates. Histopathology 2019;74:18–30. - PubMed
-
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. - PubMed
MeSH terms
Substances
Grants and funding
- 81772514/National Natural Science Foundation of China (NSFC)
- 82073381/National Natural Science Foundation of China (NSFC)
- 81725016/National Natural Science Foundation of China (NSFC)
- 81872094/National Natural Science Foundation of China (NSFC)
- 81772718/National Natural Science Foundation of China (NSFC)
- 82002684/National Natural Science Foundation of China (NSFC)
- 201806010005/Pearl River S and T Nova Program of Guangzhou Municipality (Pearl River S&T Nova Program of Guangzhou)
- S2019B151502046/Natural Science Foundation of Guangdong Province ()
- 201704020174/Natural Science Foundation of Guangdong Province ()
- 2023A1515030038/Natural Science Foundation of Guangdong Province ()
- 236464/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 2023M730374/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 2021TQ0381/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 2021M703700/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 2023BS017/Guangzhou Women and Childrens Medical Center ()
- 23KJB320018/Major Basic Research Project of the Natural Science Foundation of the Jiangsu Higher Education Institutions (Key University of Science Research Project of Jiangsu Province)
- 202201010910/Guangzhou Municipal Science and Technology Project (Guangzhou Science and Technology Plan)
- 2021TQ0381/Changzhou Institute of Technology (CIT)
- CQ20220129/Changzhou Institute of Technology (CIT)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

