An auxin research odyssey: 1989-2023
- PMID: 38382088
- PMCID: PMC11062468
- DOI: 10.1093/plcell/koae054
An auxin research odyssey: 1989-2023
Abstract
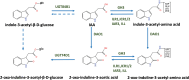
The phytohormone auxin is at times called the master regulator of plant processes and has been shown to be a central player in embryo development, the establishment of the polar axis, early aspects of seedling growth, as well as growth and organ formation during later stages of plant development. The Plant Cell has been key, since the inception of the journal, to developing an understanding of auxin biology. Auxin-regulated plant growth control is accomplished by both changes in the levels of active hormones and the sensitivity of plant tissues to these concentration changes. In this historical review, we chart auxin research as it has progressed in key areas and highlight the role The Plant Cell played in these scientific developments. We focus on understanding auxin-responsive genes, transcription factors, reporter constructs, perception, and signal transduction processes. Auxin metabolism is discussed from the development of tryptophan auxotrophic mutants, the molecular biology of conjugate formation and hydrolysis, indole-3-butyric acid metabolism and transport, and key steps in indole-3-acetic acid biosynthesis, catabolism, and transport. This progress leads to an expectation of a more comprehensive understanding of the systems biology of auxin and the spatial and temporal regulation of cellular growth and development.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures

References
-
- Abu-Zaitoon YM, Abu-Zaiton A, Tawaha ARA, Fandi KG, Alnaimat SM, Pati S, Almomani FA. Evidence from co-expression analysis for the involvement of amidase and INS in the tryptophan-independent pathway of IAA synthesis in Arabidopsis. Appl Biochem Biotechnol. 2022:194(10):4673–4682. 10.1007/s12010-022-04047-8 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

