Comprehensive analysis identifies crucial genes associated with immune cells mediating progression of carotid atherosclerotic plaque
- PMID: 38382092
- PMCID: PMC10929796
- DOI: 10.18632/aging.205566
Comprehensive analysis identifies crucial genes associated with immune cells mediating progression of carotid atherosclerotic plaque
Abstract
Backgrounds: Carotid atherosclerosis is prone to rupture and cause ischemic stroke in advanced stages of development. Our research aims to provide markers for the progression of atherosclerosis and potential targets for its treatment.
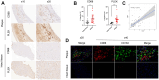
Methods: We performed a thorough analysis using various techniques including DEGs, GO/KEGG, xCell, WGCNA, GSEA, and other methods. The gene expression omnibus datasets GSE28829 and GSE43292 were utilized for this comprehensive analysis. The validation datasets employed in this study consisted of GSE41571 and GSE120521 datasets. Finally, we validated PLEK by immunohistochemistry staining in clinical samples.
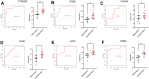
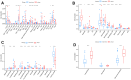
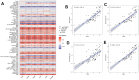
Results: Using the WGCNA technique, we discovered 636 differentially expressed genes (DEGs) and obtained 12 co-expression modules. Additionally, we discovered two modules that were specifically associated with atherosclerotic plaque. A total of 330 genes that were both present in DEGs and WGCNA results were used to create a protein-protein network in Cytoscape. We used four different algorithms to get the top 10 genes and finally got 6 overlapped genes (TYROBP, ITGB2, ITGAM, PLEK, LCP2, CD86), which are identified by GSE41571 and GSE120521 datasets. Interestingly, the area under curves (AUC) of PLEK is 0.833. Besides, we found PLEK is strongly positively correlated with most lymphocytes and myeloid cells, especially monocytes and macrophages, and negatively correlated with most stromal cells (e.g, neurons, myocytes, and fibroblasts). The expression of PLEK were consistent with the immunohistochemistry results.
Conclusions: Six genes (TYROBP, ITGB2, ITGAM, PLEK, LCP2, CD86) were found to be connected with carotid atherosclerotic plaques and PLEK may be an important biomarker and a potential therapeutic target.
Keywords: RNA sequencing; WGCNA; bioinformatics; carotid atherosclerotic plaque; immunity.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

