ATF7IP2/MCAF2 directs H3K9 methylation and meiotic gene regulation in the male germline
- PMID: 38383062
- PMCID: PMC10982687
- DOI: 10.1101/gad.351569.124
ATF7IP2/MCAF2 directs H3K9 methylation and meiotic gene regulation in the male germline
Abstract
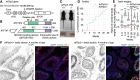
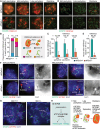
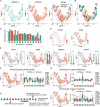
H3K9 trimethylation (H3K9me3) plays emerging roles in gene regulation, beyond its accumulation on pericentric constitutive heterochromatin. It remains a mystery why and how H3K9me3 undergoes dynamic regulation in male meiosis. Here, we identify a novel, critical regulator of H3K9 methylation and spermatogenic heterochromatin organization: the germline-specific protein ATF7IP2 (MCAF2). We show that in male meiosis, ATF7IP2 amasses on autosomal and X-pericentric heterochromatin, spreads through the entirety of the sex chromosomes, and accumulates on thousands of autosomal promoters and retrotransposon loci. On the sex chromosomes, which undergo meiotic sex chromosome inactivation (MSCI), the DNA damage response pathway recruits ATF7IP2 to X-pericentric heterochromatin, where it facilitates the recruitment of SETDB1, a histone methyltransferase that catalyzes H3K9me3. In the absence of ATF7IP2, male germ cells are arrested in meiotic prophase I. Analyses of ATF7IP2-deficient meiosis reveal the protein's essential roles in the maintenance of MSCI, suppression of retrotransposons, and global up-regulation of autosomal genes. We propose that ATF7IP2 is a downstream effector of the DDR pathway in meiosis that coordinates the organization of heterochromatin and gene regulation through the spatial regulation of SETDB1-mediated H3K9me3 deposition.
Keywords: ATF7IP2/MCAF2; H3K9me3; constitutive heterochromatin; gene activation; meiosis; meiotic sex chromosome inactivation.
© 2024 Alavattam et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




Update of
-
ATF7IP2/MCAF2 directs H3K9 methylation and meiotic gene regulation in the male germline.bioRxiv [Preprint]. 2023 Oct 2:2023.09.30.560314. doi: 10.1101/2023.09.30.560314. bioRxiv. 2023. Update in: Genes Dev. 2024 Mar 22;38(3-4):115-130. doi: 10.1101/gad.351569.124. PMID: 37873266 Free PMC article. Updated. Preprint.
References
-
- Alavattam KG, Kato Y, Sin HS, Maezawa S, Kowalski IJ, Zhang F, Pang Q, Andreassen PR, Namekawa SH. 2016. Elucidation of the Fanconi anemia protein network in meiosis and its function in the regulation of histone modifications. Cell Rep 17: 1141–1157. 10.1016/j.celrep.2016.09.073 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
