Hsa_circ_0005397 promotes hepatocellular carcinoma progression through EIF4A3
- PMID: 38383334
- PMCID: PMC10882807
- DOI: 10.1186/s12885-024-11984-6
Hsa_circ_0005397 promotes hepatocellular carcinoma progression through EIF4A3
Abstract
Purpose: The purpose of this study was to explore the expression and potential mechanism of hsa_circ_0005397 in hepatocellular carcinoma progression.
Methods: Quantitative reverse transcription-polymerase chain reaction(qRT-PCR) was used to measure the expression level of hsa_circ_0005397 and EIF4A3 from paired HCC tissues and cell lines. Western Blot (WB) and immunohistochemistry (IHC) were used to verify the protein level of EIF4A3. The specificity of primers was confirmed by agarose gel electrophoresis. Receiver Operating Characteristic (ROC) Curve was drawn to analyze diagnostic value. Actinomycin D and nuclear and cytoplasmic extraction assays were utilized to evaluate the characteristics of hsa_circ_0005397. Cell Counting kit-8 (CCK-8) and colony formation assays were performed to detect cell proliferation. Flow cytometry analysis was used to detect the cell cycle. Transwell assay was performed to determine migration and invasion ability. RNA-binding proteins (RBPs) of hsa_circ_0005397 in HCC were explored using bioinformatics websites. The relationship between hsa_circ_0005397 and Eukaryotic Translation Initiation Factor 4A3 (EIF4A3) was verified by RNA Binding Protein Immunoprecipitation (RIP) assays, correlation and rescue experiments.
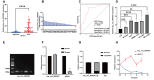
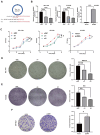
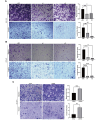
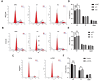
Results: In this study, hsa_circ_0005397 was found to be significantly upregulated in HCC, and the good diagnostic sensitivity and specificity shown a potential diagnostic capability. Upregulated expression of hsa_circ_0005397 was significantly related to tumor size and stage. Hsa_circ_0005397 was circular structure which more stable than liner mRNA, and mostly distributed in the cytoplasm. Upregulation of hsa_circ_0005397 generally resulted in stronger proliferative ability, clonality, and metastatic potency of HCC cells; its downregulation yielded the opposite results. EIF4A3 is an RNA-binding protein of hsa_circ_0005397, which overexpressed in paired HCC tissues and cell lines. In addition, expression of hsa_circ_0005397 decreased equally when EIF4A3 was depleted. RIP assays and correlation assay estimated that EIF4A3 could interacted with hsa_circ_0005397. Knockdown of EIF4A3 could reverse hsa_circ_0005397 function in HCC progression.
Conclusions: Hsa_circ_0005397 promotes progression of hepatocellular carcinoma through EIF4A3. These research findings may provide novel clinical value for hepatocellular carcinoma.
Keywords: EIF4A3; Hepatocellular carcinoma; hsa_circ_0005397.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
EIF4A3-Bound hsa_circ_0006847 Exerts a Tumor-Suppressive Role in Gastric Cancer.DNA Cell Biol. 2024 May;43(5):232-244. doi: 10.1089/dna.2023.0397. Epub 2024 Mar 22. DNA Cell Biol. 2024. PMID: 38513058
-
EIF4A3-Induced Upregulation of hsa_circ_0049396 Attenuates the Tumorigenesis of Nasopharyngeal Carcinoma by Regulating the Hippo-YAP Pathway.DNA Cell Biol. 2024 Oct;43(10):510-519. doi: 10.1089/dna.2024.0119. Epub 2024 Aug 12. DNA Cell Biol. 2024. PMID: 39133108
-
Circ ubiquitin-like-containing plant homeodomain and RING finger domains protein 1 increases the stability of G9a and ubiquitin-like-containing plant homeodomain and RING finger domains protein 1 messenger RNA through recruiting eukaryotic translation initiation factor 4A3, transcriptionally inhibiting PDZ and homeobox protein domain protein 1, and promotes the metastasis of hepatocellular carcinoma.J Gastroenterol Hepatol. 2024 Mar;39(3):596-607. doi: 10.1111/jgh.16408. Epub 2023 Dec 7. J Gastroenterol Hepatol. 2024. PMID: 38059880
-
Circular RNA hsa_circ_0000467 promotes colorectal cancer progression by promoting eIF4A3-mediated c-Myc translation.Mol Cancer. 2024 Jul 31;23(1):151. doi: 10.1186/s12943-024-02052-5. Mol Cancer. 2024. PMID: 39085875 Free PMC article.
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
Cited by
-
CircRAPGEF5 acts as a modulator of RAS/RAF/MEK/ERK signaling during colorectal carcinogenesis.Heliyon. 2024 Aug 10;10(16):e36133. doi: 10.1016/j.heliyon.2024.e36133. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39229520 Free PMC article.
-
Deubiquitinase USP18 mediates cell migration, apoptosis and ferroptosis in lung adenocarcinoma by depending on POU4F1/PRKAA2 axis.BMC Cancer. 2025 Mar 23;25(1):528. doi: 10.1186/s12885-025-13869-8. BMC Cancer. 2025. PMID: 40122823 Free PMC article.
-
EIF4A3-Mediated downregulation of circPTEN promotes hepatocellular carcinoma progression through the miR-1289/RBM38 Axis.J Mol Histol. 2025 Jun 17;56(4):196. doi: 10.1007/s10735-025-10473-9. J Mol Histol. 2025. PMID: 40527976
References
-
- Yang B, Zhao J, Huo T, Zhang M, Wu X. Effects of CircRNA-ITCH on proliferation and apoptosis of hepatocellular carcinoma cells through inhibiting Wnt/beta-catenin signaling pathway. J BUON. 2020;25(3):1368–74. - PubMed
-
- Cedric BC, Souraka TDM, Feng YL, Kisembo P, Tu JC. CircRNA ZFR stimulates the proliferation of hepatocellular carcinoma through upregulating MAP2K1. Eur Rev Med Pharmacol Sci. 2020;24(19):9924–31. - PubMed
-
- Guo XL, Wang HB, Yong JK, Zhong J, Li QH. MiR-128-3p overexpression sensitizes hepatocellular carcinoma cells to sorafenib induced apoptosis through regulating DJ-1. Eur Rev Med Pharmacol Sci. 2018;22(20):6667–77. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

