Correlative light-electron microscopy methods to characterize the ultrastructural features of the replicative and dormant liver stages of Plasmodium parasites
- PMID: 38383417
- PMCID: PMC10882739
- DOI: 10.1186/s12936-024-04862-w
Correlative light-electron microscopy methods to characterize the ultrastructural features of the replicative and dormant liver stages of Plasmodium parasites
Abstract
Background: The infection of the liver by Plasmodium parasites is an obligatory step leading to malaria disease. Following hepatocyte invasion, parasites differentiate into replicative liver stage schizonts and, in the case of Plasmodium species causing relapsing malaria, into hypnozoites that can lie dormant for extended periods of time before activating. The liver stages of Plasmodium remain elusive because of technical challenges, including low infection rate. This has been hindering experimentations with well-established technologies, such as electron microscopy. A deeper understanding of hypnozoite biology could prove essential in the development of radical cure therapeutics against malaria.
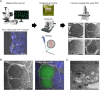
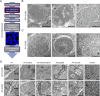
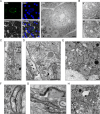
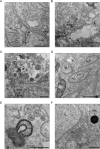
Results: The liver stages of the rodent parasite Plasmodium berghei, causing non-relapsing malaria, and the simian parasite Plasmodium cynomolgi, causing relapsing malaria, were characterized in human Huh7 cells or primary non-human primate hepatocytes using Correlative Light-Electron Microscopy (CLEM). Specifically, CLEM approaches that rely on GFP-expressing parasites (GFP-CLEM) or on an immunofluorescence assay (IFA-CLEM) were used for imaging liver stages. The results from P. berghei showed that host and parasite organelles can be identified and imaged at high resolution using both CLEM approaches. While IFA-CLEM was associated with more pronounced extraction of cellular content, samples' features were generally well preserved. Using IFA-CLEM, a collection of micrographs was acquired for P. cynomolgi liver stage schizonts and hypnozoites, demonstrating the potential of this approach for characterizing the liver stages of Plasmodium species causing relapsing malaria.
Conclusions: A CLEM approach that does not rely on parasites expressing genetically encoded tags was developed, therefore suitable for imaging the liver stages of Plasmodium species that lack established protocols to perform genetic engineering. This study also provides a dataset that characterizes the ultrastructural features of liver stage schizonts and hypnozoites from the simian parasite species P. cynomolgi.
Keywords: CLEM; Hepatocytes; Hypnozoites; Mitochondria; Plasmodium berghei; Plasmodium cynomolgi; Relapsing malaria; Schizonts; TEM; Transmission electron microscopy.
© 2024. The Author(s).
Conflict of interest statement
GM, LT, MEF, ML, VC, ELF, AH and SAM were employed by and/or shareholders of Novartis Pharma AG during this study.
Figures

References
-
- WHO . World malaria report. Geneva: World Health Organization; 2023.

