Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci
- PMID: 38383453
- PMCID: PMC10881467
- DOI: 10.1038/s41467-024-45381-1
Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci
Erratum in
-
Author Correction: Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci.Nat Commun. 2024 May 10;15(1):3935. doi: 10.1038/s41467-024-48497-6. Nat Commun. 2024. PMID: 38729949 Free PMC article. No abstract available.
Abstract
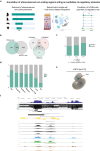
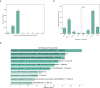
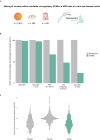
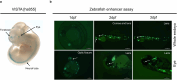
Cross-species genome comparisons have revealed a substantial number of ultraconserved non-coding elements (UCNEs). Several of these elements have proved to be essential tissue- and cell type-specific cis-regulators of developmental gene expression. Here, we characterize a set of UCNEs as candidate CREs (cCREs) during retinal development and evaluate the contribution of their genomic variation to rare eye diseases, for which pathogenic non-coding variants are emerging. Integration of bulk and single-cell retinal multi-omics data reveals 594 genes under potential cis-regulatory control of UCNEs, of which 45 are implicated in rare eye disease. Mining of candidate cis-regulatory UCNEs in WGS data derived from the rare eye disease cohort of Genomics England reveals 178 ultrarare variants within 84 UCNEs associated with 29 disease genes. Overall, we provide a comprehensive annotation of ultraconserved non-coding regions acting as cCREs during retinal development which can be targets of non-coding variation underlying rare eye diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Comparative 3D genome analysis between neural retina and retinal pigment epithelium reveals differential cis-regulatory interactions at retinal disease loci.Genome Biol. 2024 May 17;25(1):123. doi: 10.1186/s13059-024-03250-6. Genome Biol. 2024. PMID: 38760655 Free PMC article.
-
Multi-omics approach dissects cis-regulatory mechanisms underlying North Carolina macular dystrophy, a retinal enhanceropathy.Am J Hum Genet. 2022 Nov 3;109(11):2029-2048. doi: 10.1016/j.ajhg.2022.09.013. Epub 2022 Oct 14. Am J Hum Genet. 2022. PMID: 36243009 Free PMC article.
-
Genomic context analysis reveals dense interaction network between vertebrate ultraconserved non-coding elements.Bioinformatics. 2012 Sep 15;28(18):i395-i401. doi: 10.1093/bioinformatics/bts400. Bioinformatics. 2012. PMID: 22962458 Free PMC article.
-
High-Throughput Analysis of Retinal Cis-Regulatory Networks by Massively Parallel Reporter Assays.Adv Exp Med Biol. 2019;1185:359-364. doi: 10.1007/978-3-030-27378-1_59. Adv Exp Med Biol. 2019. PMID: 31884638 Free PMC article. Review.
-
Cis-regulatory mutations in human disease.Brief Funct Genomic Proteomic. 2009 Jul;8(4):310-6. doi: 10.1093/bfgp/elp021. Epub 2009 Jul 29. Brief Funct Genomic Proteomic. 2009. PMID: 19641089 Free PMC article. Review.
Cited by
-
Decoding physiological and pathological roles of innate immune cells in eye diseases: the perspectives from single-cell RNA sequencing.Front Immunol. 2024 Oct 31;15:1490719. doi: 10.3389/fimmu.2024.1490719. eCollection 2024. Front Immunol. 2024. PMID: 39544948 Free PMC article. Review.
-
Multi Omics Applications in Biological Systems.Curr Issues Mol Biol. 2024 Jun 11;46(6):5777-5793. doi: 10.3390/cimb46060345. Curr Issues Mol Biol. 2024. PMID: 38921016 Free PMC article. Review.
-
Epigenome-metabolism nexus in the retina: implications for aging and disease.Trends Genet. 2024 Aug;40(8):718-729. doi: 10.1016/j.tig.2024.04.012. Epub 2024 May 22. Trends Genet. 2024. PMID: 38782642 Free PMC article. Review.
-
The Human Cell Atlas from a cell census to a unified foundation model.Nature. 2025 Jan;637(8048):1065-1071. doi: 10.1038/s41586-024-08338-4. Epub 2024 Nov 20. Nature. 2025. PMID: 39566552
-
Brain-heart-eye axis revealed by multi-organ imaging, genetics and proteomics.medRxiv [Preprint]. 2025 Jun 9:2025.01.04.25319995. doi: 10.1101/2025.01.04.25319995. medRxiv. 2025. PMID: 40585072 Free PMC article. Preprint.
References
MeSH terms
Grants and funding
- 1802220N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- 813490/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- 813490/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)

