HMGA1 drives chemoresistance in esophageal squamous cell carcinoma by suppressing ferroptosis
- PMID: 38383528
- PMCID: PMC10881472
- DOI: 10.1038/s41419-024-06467-2
HMGA1 drives chemoresistance in esophageal squamous cell carcinoma by suppressing ferroptosis
Abstract
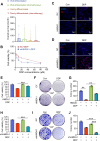
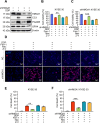
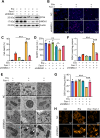
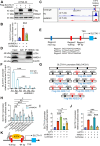
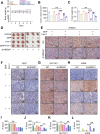
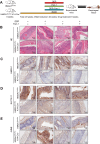
Chemotherapy is a primary treatment for esophageal squamous cell carcinoma (ESCC). Resistance to chemotherapeutic drugs is an important hurdle to effective treatment. Understanding the mechanisms underlying chemotherapy resistance in ESCC is an unmet medical need to improve the survival of ESCC. Herein, we demonstrate that ferroptosis triggered by inhibiting high mobility group AT-hook 1 (HMGA1) may provide a novel opportunity to gain an effective therapeutic strategy against chemoresistance in ESCC. HMGA1 is upregulated in ESCC and works as a key driver for cisplatin (DDP) resistance in ESCC by repressing ferroptosis. Inhibition of HMGA1 enhances the sensitivity of ESCC to ferroptosis. With a transcriptome analysis and following-up assays, we demonstrated that HMGA1 upregulates the expression of solute carrier family 7 member 11 (SLC7A11), a key transporter maintaining intracellular glutathione homeostasis and inhibiting the accumulation of malondialdehyde (MDA), thereby suppressing cell ferroptosis. HMGA1 acts as a chromatin remodeling factor promoting the binding of activating transcription factor 4 (ATF4) to the promoter of SLC7A11, and hence enhancing the transcription of SLC7A11 and maintaining the redox balance. We characterized that the enhanced chemosensitivity of ESCC is primarily attributed to the increased susceptibility of ferroptosis resulting from the depletion of HMGA1. Moreover, we utilized syngeneic allograft tumor models and genetically engineered mice of HMGA1 to induce ESCC and validated that depletion of HMGA1 promotes ferroptosis and restores the sensitivity of ESCC to DDP, and hence enhances the therapeutic efficacy. Our finding uncovers a critical role of HMGA1 in the repression of ferroptosis and thus in the establishment of DDP resistance in ESCC, highlighting HMGA1-based rewiring strategies as potential approaches to overcome ESCC chemotherapy resistance. Schematic depicting that HMGA1 maintains intracellular redox homeostasis against ferroptosis by assisting ATF4 to activate SLC7A11 transcription, resulting in ESCC resistance to chemotherapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

