Increased CO2 fixation enables high carbon-yield production of 3-hydroxypropionic acid in yeast
- PMID: 38383540
- PMCID: PMC10881976
- DOI: 10.1038/s41467-024-45557-9
Increased CO2 fixation enables high carbon-yield production of 3-hydroxypropionic acid in yeast
Abstract
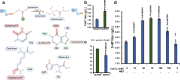
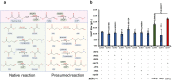
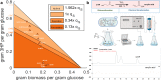
CO2 fixation plays a key role to make biobased production cost competitive. Here, we use 3-hydroxypropionic acid (3-HP) to showcase how CO2 fixation enables approaching theoretical-yield production. Using genome-scale metabolic models to calculate the production envelope, we demonstrate that the provision of bicarbonate, formed from CO2, restricts previous attempts for high yield production of 3-HP. We thus develop multiple strategies for bicarbonate uptake, including the identification of Sul1 as a potential bicarbonate transporter, domain swapping of malonyl-CoA reductase, identification of Esbp6 as a potential 3-HP exporter, and deletion of Uga1 to prevent 3-HP degradation. The combined rational engineering increases 3-HP production from 0.14 g/L to 11.25 g/L in shake flask using 20 g/L glucose, approaching the maximum theoretical yield with concurrent biomass formation. The engineered yeast forms the basis for commercialization of bio-acrylic acid, while our CO2 fixation strategies pave the way for CO2 being used as the sole carbon source.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hertel TW, et al. Effects of us maize ethanol on global land use and greenhouse gas emissions: estimating market-mediated responses. Bioscience. 2010;60:223–231. doi: 10.1525/bio.2010.60.3.8. - DOI
-
- Liu Z, Wang K, Chen Y, Tan T, Nielsen J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 2020;3:274–CO288. doi: 10.1038/s41929-019-0421-5. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

