Integrating single-cell and spatially resolved transcriptomic strategies to survey the astrocyte response to stroke in male mice
- PMID: 38383565
- PMCID: PMC10882052
- DOI: 10.1038/s41467-024-45821-y
Integrating single-cell and spatially resolved transcriptomic strategies to survey the astrocyte response to stroke in male mice
Abstract
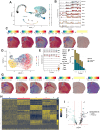
Astrocytes, a type of glial cell in the central nervous system (CNS), adopt diverse states in response to injury that are influenced by their location relative to the insult. Here, we describe a platform for spatially resolved, single-cell transcriptomics and proteomics, called tDISCO (tissue-digital microfluidic isolation of single cells for -Omics). We use tDISCO alongside two high-throughput platforms for spatial (Visium) and single-cell transcriptomics (10X Chromium) to examine the heterogeneity of the astrocyte response to a cortical ischemic stroke in male mice. We show that integration of Visium and 10X Chromium datasets infers two astrocyte populations, proximal or distal to the injury site, while tDISCO determines the spatial boundaries and molecular profiles that define these populations. We find that proximal astrocytes show differences in lipid shuttling, with enriched expression of Apoe and Fabp5. Our datasets provide a resource for understanding the roles of astrocytes in stroke and showcase the utility of tDISCO for hypothesis-driven, spatially resolved single-cell experiments.
© 2024. The Author(s).
Conflict of interest statement
M.D. and A.R.W. are co-inventors on a patent application (Applicants: The governing council of the University of Toronto, and Sinai Health System; Inventors: Michael Dryden and Aaron R Wheeler; Application Number: PCT/CA2017/051158 (filed 20.09.2019); Status: Pending; Specific Aspect of Manuscript: System and method for identifying and targeting individual cells within a cell population for selective extraction of cellular content with a pulsed laser within a digital microfluidic device.
Figures




References
-
- Palla G., Fischer D. S., Regev A., Theis F. J. Spatial components of molecular tissue biology. 10.1038/s41587-021-01182-1. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

