Data encoding for healthcare data democratization and information leakage prevention
- PMID: 38383571
- PMCID: PMC10882022
- DOI: 10.1038/s41467-024-45777-z
Data encoding for healthcare data democratization and information leakage prevention
Abstract
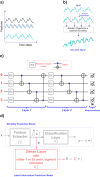
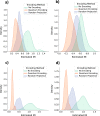
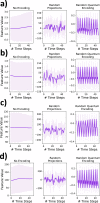
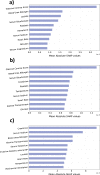
The lack of data democratization and information leakage from trained models hinder the development and acceptance of robust deep learning-based healthcare solutions. This paper argues that irreversible data encoding can provide an effective solution to achieve data democratization without violating the privacy constraints imposed on healthcare data and clinical models. An ideal encoding framework transforms the data into a new space where it is imperceptible to a manual or computational inspection. However, encoded data should preserve the semantics of the original data such that deep learning models can be trained effectively. This paper hypothesizes the characteristics of the desired encoding framework and then exploits random projections and random quantum encoding to realize this framework for dense and longitudinal or time-series data. Experimental evaluation highlights that models trained on encoded time-series data effectively uphold the information bottleneck principle and hence, exhibit lesser information leakage from trained models.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Goodfellow, I., Bengio, Y. & Courville, A. Deep learning. http://www.deeplearningbook.org (MIT Press, 2016).
Grants and funding
LinkOut - more resources
Full Text Sources

