Intestinal IL-22RA1 signaling regulates intrinsic and systemic lipid and glucose metabolism to alleviate obesity-associated disorders
- PMID: 38383607
- PMCID: PMC10881576
- DOI: 10.1038/s41467-024-45568-6
Intestinal IL-22RA1 signaling regulates intrinsic and systemic lipid and glucose metabolism to alleviate obesity-associated disorders
Abstract
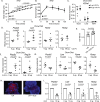
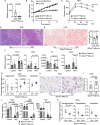
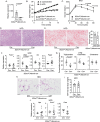
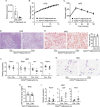
IL-22 is critical for ameliorating obesity-induced metabolic disorders. However, it is unknown where IL-22 acts to mediate these outcomes. Here we examine the importance of tissue-specific IL-22RA1 signaling in mediating long-term high fat diet (HFD) driven metabolic disorders. To do so, we generated intestinal epithelium-, liver-, and white adipose tissue (WAT)-specific Il22ra1 knockout and littermate control mice. Intestinal epithelium- and liver-specific IL-22RA1 signaling upregulated systemic glucose metabolism. Intestinal IL-22RA1 signaling also mediated liver and WAT metabolism in a microbiota-dependent manner. We identified an association between Oscillibacter and elevated WAT inflammation, likely induced by Mmp12 expressing macrophages. Mechanistically, transcription of intestinal lipid metabolism genes is regulated by IL-22 and potentially IL-22-induced IL-18. Lastly, we show that Paneth cell-specific IL-22RA1 signaling, in part, mediates systemic glucose metabolism after HFD. Overall, these results elucidate a key role of intestinal epithelium-specific IL-22RA1 signaling in regulating intestinal metabolism and alleviating systemic obesity-associated disorders.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

