Heterogeneity of synaptic connectivity in the fly visual system
- PMID: 38383614
- PMCID: PMC10882054
- DOI: 10.1038/s41467-024-45971-z
Heterogeneity of synaptic connectivity in the fly visual system
Abstract
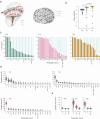
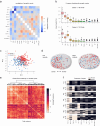
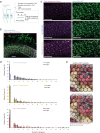
Visual systems are homogeneous structures, where repeating columnar units retinotopically cover the visual field. Each of these columns contain many of the same neuron types that are distinguished by anatomic, genetic and - generally - by functional properties. However, there are exceptions to this rule. In the 800 columns of the Drosophila eye, there is an anatomically and genetically identifiable cell type with variable functional properties, Tm9. Since anatomical connectivity shapes functional neuronal properties, we identified the presynaptic inputs of several hundred Tm9s across both optic lobes using the full adult female fly brain (FAFB) electron microscopic dataset and FlyWire connectome. Our work shows that Tm9 has three major and many sparsely distributed inputs. This differs from the presynaptic connectivity of other Tm neurons, which have only one major, and more stereotypic inputs than Tm9. Genetic synapse labeling showed that the heterogeneous wiring exists across individuals. Together, our data argue that the visual system uses heterogeneous, distributed circuit properties to achieve robust visual processing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Fischbach, K.-F & Dittrich, A. P. M. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 (1989).
MeSH terms
Grants and funding
- n.a./Studienstiftung des Deutschen Volkes (German National Academic Foundation)
- U24 NS126935/NS/NINDS NIH HHS/United States
- 716512/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- GRK2526/1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- RF1 MH117815/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

