Haplotype information of large neuromuscular disease genes provided by linked-read sequencing has a potential to increase diagnostic yield
- PMID: 38383731
- PMCID: PMC10881483
- DOI: 10.1038/s41598-024-54866-4
Haplotype information of large neuromuscular disease genes provided by linked-read sequencing has a potential to increase diagnostic yield
Abstract
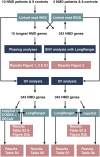
Rare or novel missense variants in large genes such as TTN and NEB are frequent in the general population, which hampers the interpretation of putative disease-causing biallelic variants in patients with sporadic neuromuscular disorders. Often, when the first initial genetic analysis is performed, the reconstructed haplotype, i.e. phasing information of the variants is missing. Segregation analysis increases the diagnostic turnaround time and is not always possible if samples from family members are lacking. To overcome this difficulty, we investigated how well the linked-read technology succeeded to phase variants in these large genes, and whether it improved the identification of structural variants. Linked-read sequencing data of nemaline myopathy, distal myopathy, and proximal myopathy patients were analyzed for phasing, single nucleotide variants, and structural variants. Variant phasing was successful in the large muscle genes studied. The longest continuous phase blocks were gained using high-quality DNA samples with long DNA fragments. Homozygosity increased the number of phase blocks, especially in exome sequencing samples lacking intronic variation. In our cohort, linked-read sequencing added more information about the structural variation but did not lead to a molecular genetic diagnosis. The linked-read technology can support the clinical diagnosis of neuromuscular and other genetic disorders.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

