Leveraging electronic health records and knowledge networks for Alzheimer's disease prediction and sex-specific biological insights
- PMID: 38383858
- PMCID: PMC10950787
- DOI: 10.1038/s43587-024-00573-8
Leveraging electronic health records and knowledge networks for Alzheimer's disease prediction and sex-specific biological insights
Abstract
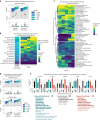
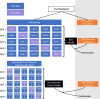
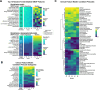
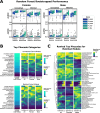
Identification of Alzheimer's disease (AD) onset risk can facilitate interventions before irreversible disease progression. We demonstrate that electronic health records from the University of California, San Francisco, followed by knowledge networks (for example, SPOKE) allow for (1) prediction of AD onset and (2) prioritization of biological hypotheses, and (3) contextualization of sex dimorphism. We trained random forest models and predicted AD onset on a cohort of 749 individuals with AD and 250,545 controls with a mean area under the receiver operating characteristic of 0.72 (7 years prior) to 0.81 (1 day prior). We further harnessed matched cohort models to identify conditions with predictive power before AD onset. Knowledge networks highlight shared genes between multiple top predictors and AD (for example, APOE, ACTB, IL6 and INS). Genetic colocalization analysis supports AD association with hyperlipidemia at the APOE locus, as well as a stronger female AD association with osteoporosis at a locus near MS4A6A. We therefore show how clinical data can be utilized for early AD prediction and identification of personalized biological hypotheses.
© 2024. The Author(s).
Conflict of interest statement
Unrelated to the work described in this manuscript, R.B. has received research support from F Hoffmann-La Roche, Novartis and Biogen and has received personal support for consulting and/or scientific advisory boards from Alexion, EMD Serono, Horizon, Jansen and TG Therapeutics. Also unrelated to the work, K.P.R. has served on a medical advisory board for Eli Lily. S.B. is co-founder of Mate Bioservices. J.R. has previously interned at Roche. The remaining authors declare no competing interests.
Figures









References
-
- 2022 Alzheimer’s disease facts and figures. Alzheimers Dement. 18, 700–789 (2022). - PubMed
-
- Savonenko, A. V., Wong, P. C., & Li, T. Alzheimer diseases. In Neurobiology of Brain Disorders: Biological Basis of Neurological and Psychiatric Disorders, 2nd Edition (eds Zigmond, M. .J. et al.) 313–336 (Elsevier, 2023). 10.1016/b978-0-323-85654-6.00022-8
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

