Hyperacetylated histone H4 is a source of carbon contributing to lipid synthesis
- PMID: 38383863
- PMCID: PMC10987603
- DOI: 10.1038/s44318-024-00053-0
Hyperacetylated histone H4 is a source of carbon contributing to lipid synthesis
Abstract
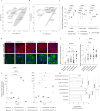
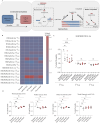
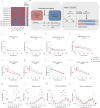
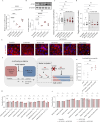
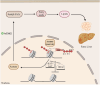
Histone modifications commonly integrate environmental cues with cellular metabolic outputs by affecting gene expression. However, chromatin modifications such as acetylation do not always correlate with transcription, pointing towards an alternative role of histone modifications in cellular metabolism. Using an approach that integrates mass spectrometry-based histone modification mapping and metabolomics with stable isotope tracers, we demonstrate that elevated lipids in acetyltransferase-depleted hepatocytes result from carbon atoms derived from deacetylation of hyperacetylated histone H4 flowing towards fatty acids. Consistently, enhanced lipid synthesis in acetyltransferase-depleted hepatocytes is dependent on histone deacetylases and acetyl-CoA synthetase ACSS2, but not on the substrate specificity of the acetyltransferases. Furthermore, we show that during diet-induced lipid synthesis the levels of hyperacetylated histone H4 decrease in hepatocytes and in mouse liver. In addition, overexpression of acetyltransferases can reverse diet-induced lipogenesis by blocking lipid droplet accumulation and maintaining the levels of hyperacetylated histone H4. Overall, these findings highlight hyperacetylated histones as a metabolite reservoir that can directly contribute carbon to lipid synthesis, constituting a novel function of chromatin in cellular metabolism.
Keywords: Acetylation; Epigenetics; Histone Reservoirs; Lipid Metabolism.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
- 890750/EC | Horizon Europe | Excellent Science | HORIZON EUROPE Marie Sklodowska-Curie Actions (MSCA)
- 0000424/European Proteomics Infrastructure Consortium providing access (EPIC-XS)
- 0000463/European Proteomics Infrastructure Consortium providing access (EPIC-XS)
- EXCELLENCE/0421/0342/Research and Innovation Foundation (RIF)
- EXCELLENCE/0421/0302/Research and Innovation Foundation (RIF)
LinkOut - more resources
Full Text Sources
Research Materials

