PRMT1 Sustains De Novo Fatty Acid Synthesis by Methylating PHGDH to Drive Chemoresistance in Triple-Negative Breast Cancer
- PMID: 38383964
- PMCID: PMC10982647
- DOI: 10.1158/0008-5472.CAN-23-2266
PRMT1 Sustains De Novo Fatty Acid Synthesis by Methylating PHGDH to Drive Chemoresistance in Triple-Negative Breast Cancer
Abstract
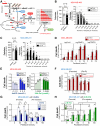
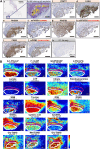
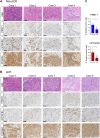
Triple-negative breast cancer (TNBC) chemoresistance hampers the ability to effectively treat patients. Identification of mechanisms driving chemoresistance can lead to strategies to improve treatment. Here, we revealed that protein arginine methyltransferase-1 (PRMT1) simultaneously methylates D-3-phosphoglycerate dehydrogenase (PHGDH), a critical enzyme in serine synthesis, and the glycolytic enzymes PFKFB3 and PKM2 in TNBC cells. 13C metabolic flux analyses showed that PRMT1-dependent methylation of these three enzymes diverts glucose toward intermediates in the serine-synthesizing and serine/glycine cleavage pathways, thereby accelerating the production of methyl donors in TNBC cells. Mechanistically, PRMT1-dependent methylation of PHGDH at R54 or R20 activated its enzymatic activity by stabilizing 3-phosphoglycerate binding and suppressing polyubiquitination. PRMT1-mediated PHGDH methylation drove chemoresistance independently of glutathione synthesis. Rather, activation of the serine synthesis pathway supplied α-ketoglutarate and citrate to increase palmitate levels through activation of fatty acid synthase (FASN). Increased palmitate induced protein S-palmitoylation of PHGDH and FASN to further enhance fatty acid synthesis in a PRMT1-dependent manner. Loss of PRMT1 or pharmacologic inhibition of FASN or protein S-palmitoyltransferase reversed chemoresistance in TNBC. Furthermore, IHC coupled with imaging MS in clinical TNBC specimens substantiated that PRMT1-mediated methylation of PHGDH, PFKFB3, and PKM2 correlates with chemoresistance and that metabolites required for methylation and fatty acid synthesis are enriched in TNBC. Together, these results suggest that enhanced de novo fatty acid synthesis mediated by coordinated protein arginine methylation and protein S-palmitoylation is a therapeutic target for overcoming chemoresistance in TNBC.
Significance: PRMT1 promotes chemoresistance in TNBC by methylating metabolic enzymes PFKFB3, PKM2, and PHGDH to augment de novo fatty acid synthesis, indicating that targeting this axis is a potential treatment strategy.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Li S, Wu H, Huang X, Jian Y, Kong L, Xu H, et al. . BOP1 confers chemoresistance of triple-negative breast cancer by promoting CBP-mediated β-catenin acetylation. J Pathol 2021;254:265–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 21K06072(C)/Japan Society for the Promotion of Science (JSPS)
- 15H04355(B)/Japan Society for the Promotion of Science (JSPS)
- 17K19614/Japan Society for the Promotion of Science (JSPS)
- JPMJER0906/Exploratory Research for Advanced Technology (ERATO)
- JP22zf0127007/Moonshot Research and Development Program (Moonshot)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

