ISG15/USP18/STAT2 is a molecular hub regulating IFN I-mediated control of Dengue and Zika virus replication
- PMID: 38384473
- PMCID: PMC10879325
- DOI: 10.3389/fimmu.2024.1331731
ISG15/USP18/STAT2 is a molecular hub regulating IFN I-mediated control of Dengue and Zika virus replication
Abstract
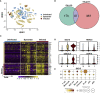
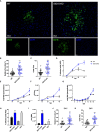
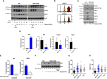
The establishment of a virus infection is the result of the pathogen's ability to replicate in a hostile environment generated by the host's immune system. Here, we found that ISG15 restricts Dengue and Zika viruses' replication through the stabilization of its binding partner USP18. ISG15 expression was necessary to control DV replication driven by both autocrine and paracrine type one interferon (IFN-I) signaling. Moreover, USP18 competes with NS5-mediated STAT2 degradation, a major mechanism for establishment of flavivirus infection. Strikingly, reconstitution of USP18 in ISG15-deficient cells was sufficient to restore the STAT2's stability and restrict virus growth, suggesting that the IFNAR-mediated ISG15 activity is also antiviral. Our results add a novel layer of complexity in the virus/host interaction interface and suggest that NS5 has a narrow window of opportunity to degrade STAT2, therefore suppressing host's IFN-I mediated response and promoting virus replication.
Keywords: Dengue virus; ISG15; ISGylation; USP18; Zika virus; antiviral response; immune evasion; type one interferon.
Copyright © 2024 Espada, da Rocha, Ricciardi-Jorge, dos Santos, Soares, Malaquias, Patrício, Gonzalez Kozlova, dos Santos, Bordignon, Sanford, Fajardo, Sweeney, Báfica and Mansur.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

