Revisiting genomes of non-model species with long reads yields new insights into their biology and evolution
- PMID: 38384712
- PMCID: PMC10879605
- DOI: 10.3389/fgene.2024.1308527
Revisiting genomes of non-model species with long reads yields new insights into their biology and evolution
Abstract
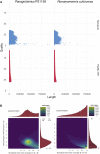
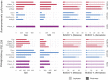
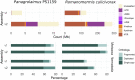
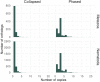
High-quality genomes obtained using long-read data allow not only for a better understanding of heterozygosity levels, repeat content, and more accurate gene annotation and prediction when compared to those obtained with short-read technologies, but also allow to understand haplotype divergence. Advances in long-read sequencing technologies in the last years have made it possible to produce such high-quality assemblies for non-model organisms. This allows us to revisit genomes, which have been problematic to scaffold to chromosome-scale with previous generations of data and assembly software. Nematoda, one of the most diverse and speciose animal phyla within metazoans, remains poorly studied, and many previously assembled genomes are fragmented. Using long reads obtained with Nanopore R10.4.1 and PacBio HiFi, we generated highly contiguous assemblies of a diploid nematode of the Mermithidae family, for which no closely related genomes are available to date, as well as a collapsed assembly and a phased assembly for a triploid nematode from the Panagrolaimidae family. Both genomes had been analysed before, but the fragmented assemblies had scaffold sizes comparable to the length of long reads prior to assembly. Our new assemblies illustrate how long-read technologies allow for a much better representation of species genomes. We are now able to conduct more accurate downstream assays based on more complete gene and transposable element predictions.
Keywords: Nematoda; genome annotation; genome assembly; genomics; long reads.
Copyright © 2024 Guiglielmoni, Villegas, Kirangwa and Schiffer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing.BMC Genomics. 2019 Jan 9;20(1):23. doi: 10.1186/s12864-018-5381-7. BMC Genomics. 2019. PMID: 30626323 Free PMC article.
-
Completion of draft bacterial genomes by long-read sequencing of synthetic genomic pools.BMC Genomics. 2020 Jul 29;21(1):519. doi: 10.1186/s12864-020-06910-6. BMC Genomics. 2020. PMID: 32727443 Free PMC article.
-
The long and short of it: benchmarking viromics using Illumina, Nanopore and PacBio sequencing technologies.Microb Genom. 2024 Feb;10(2):001198. doi: 10.1099/mgen.0.001198. Microb Genom. 2024. PMID: 38376377 Free PMC article.
-
Complex genome assembly based on long-read sequencing.Brief Bioinform. 2022 Sep 20;23(5):bbac305. doi: 10.1093/bib/bbac305. Brief Bioinform. 2022. PMID: 35940845 Review.
-
Genome Sequencing and Assembly by Long Reads in Plants.Genes (Basel). 2017 Dec 28;9(1):6. doi: 10.3390/genes9010006. Genes (Basel). 2017. PMID: 29283420 Free PMC article. Review.
Cited by
-
Insights from draft genomes of Heterodera species isolated from field soil samples.BMC Genomics. 2025 Feb 18;26(1):158. doi: 10.1186/s12864-025-11351-0. BMC Genomics. 2025. PMID: 39966714 Free PMC article.
-
Armless hairpin-like tRNAs in Romanomermis culicivorax: Evolutionary adaptation of a mitochondrial elongation factor EF-Tu.J Biol Chem. 2025 Jul;301(7):110294. doi: 10.1016/j.jbc.2025.110294. Epub 2025 May 24. J Biol Chem. 2025. PMID: 40419126 Free PMC article.
-
A reference genome enhances the power to detect signatures of recent anthropogenic impact in genomic data: a lesson learned from a stag beetle system.BMC Biol. 2025 Jul 9;23(1):205. doi: 10.1186/s12915-025-02307-7. BMC Biol. 2025. PMID: 40629430 Free PMC article.
-
An alternative adaptation strategy of the CCA-adding enzyme to accept noncanonical tRNA substrates in Ascaris suum.J Biol Chem. 2025 Apr;301(4):108414. doi: 10.1016/j.jbc.2025.108414. Epub 2025 Mar 17. J Biol Chem. 2025. PMID: 40107618 Free PMC article.
References
LinkOut - more resources
Full Text Sources

